2.College of Life Sciences, Northeast Forestry University, Harbin, 150040, P.R. China
3.College of Agriculture, Northeast Agricultural University, Harbin, 150090, P.R. China
 Author
Author  Correspondence author
Correspondence author
Computational Molecular Biology, 2011, Vol. 1, No. 2 doi: 10.5376/cmb.2011.01.0002
Received: 14 Oct., 2011 Accepted: 26 Nov., 2011 Published: 29 Dec., 2011
Wang et al., 2011, Pathogenesis Related Protein (PR Protein) in Soybean Predicted Through HMMER and BLAST Resources, Genomics and Applied Biology, 30(6): 649-656 (doi:10.3969/gab.030.000649)
The production and accumulation of PR (pathogenesis related protein) protein in plant are the main characteristics in the responses of biotic and abiotic stress. In recent years a large number of PR proteins have been identified, which were divided into 14 functional families based on their structure, phylogenetic and biological activities. However, little PR protein has been found in soybean and cereal grain crops. In this paper we acquired 36 PR protein members of 9 families predicted through the BLAST and HMMER program with the queries for all the PR proteins in Arabidopsis, rice, corn and legumes. A comprehensive analysis has been carried out by the aspects of the PR gene distribution, gene structure, length, number of extron, and evolutionary relationships. The predicted PR proteins in this paper might provide a good foundation for disease resistance in soybean breeding program and disease resistance genetic engineering, as well as provide a powerful gene prediction approach for other gene family in soybean genetics research.
There are several new proteins which were present in many species of plants infected with the pathogen or induced by some specific compounds, all of these proteins have been found infected with the pathogen later, and known as pathogenesis-related protein. They have the ability of anti-fungal or bacterial, when a large number of these proteins were produced in the infection site, forming a protective barrier against pathogens to reduce the sensitivity of plants (Edreva, 2005). PR protein was detected when the tobacco mosaic virus (TMV) infected tobacco leaves initially, firstly called the b-protein, and then renamed pathogenesis-related proteins (van Loon and van Kammen, 1970). The PR proteins of the same family had higher homology sequences and the similar function, contrariwise, they have different functions, and most of them were enzymes, such as chitinase (Wen et al., 2008). PR protein was originally divided into five groups (PR-1 to PR-5), in the research of tobacco. They were classified by molecular genetic techniques, sorted according to the electrophoretic mobility. Each member in one group has a similar composition (Bol et al., 1990). PR-1 group, the most abundant, reached 1%~2% of total leaf protein. PR-5 group was thaumatin-like protein (TLP), which could biodegradable fungal cell membrane, especially had a strong resistance to Oomycetes (Batalia et al., 1996). And it can activate the activity of the resistant protein to serine endopeptidase enzyme.
According to structural features, the PR protein can be divided into 14 families (Table 1) (van Loon et al., 1994; van Loon and van Strien, 1999).
.png) Table 1 Recognized and proposed families of pathogenesis-related proteins |
However, as the further research and its improvement, the PR proteins were divided into 17 families (Wang, 1995), in which PR15 and PR16 were similar to germination or germin-like protein. At present, only five PR proteins in soybean were identified. While only PR1 and thaumatin-like protein were reported in the references (Graham, 2005). Therefore, it has a very important significance to predict and investigate PR protein of soybean.
In this paper, we predicted the candidate PR protein sequence in soybean by BLAST and HMMER methods based on assembled the PR protein sequences from different species. And a detailed analysis was made on the linkage group of PR proteins and their distribution, gene structure, gene length, and the relationship between the evolutions.
1 Results and Analysis
1.1 Obtain candidate PR protein sequences by BLAST
A lot of PR proteins homologous sequences were obtained by BLAST method, such as in PR-1 family, we gained 79 PR proteins and found a protein sequence with a typical domain by multiple sequence alignments. Figure 1 showed the conserved domain of PR1 partial protein via sequence alignment.
.png) Figure 1 Sequence alignment of partial PR1 protein |
More detailed genetic information was obtained by the method of BLAST. Table 2 showed the PR proteins which were obtained by BLAST, assembling, and prediction. In PR1 and PR5 family, both of them were predicted six homologous PR proteins, while in the original database, PR13 and PR14 family were discarded, because either there were not enough sequences or the lower score in homology match.
Table2 Sequence and accession number of candidate PR protein |
1.2 Obtain the CDS of candidate PR protein sequences by HMMER
Only CDS of the candidate PR protein sequences can be obtained, because the database was established with the CDS of the genome. The linkage group distribution and length of CDS can be obtained by the HMMER program (Table 3).
 Table 3 CDS of candidate PR protein sequences by HMMER |
1.3 The prediction and analysis of soybean PR protein sequences
The sequences from the two prediction methods were alignment, and the repetitive sequences were removed. The unique sequences were assembled and extended. And then we submitted the assembling sequences to NCBI for annotation, at last, we determined the real CDS sequences. Among them, the members of PR1, PR3, PR5, PR6, PR10, and PRNF all have reduced, PR4 family has no repetitive, while PR2 and PR12 families have been removed because both the sequences of prediction were just the duplication of other families, and these two families were not shown in the later results.
The character of PR proteins with low molecular weight (6 ~ 43 kD) was stable when the pH value was lower than 3.0, and it has a higher resistance to the protease, so the PR proteins are established in all plant organs – leaves, stems, roots, flowers, particularly abundant in the leaves. PRs have dual cellular localization – vacuolar and apoplastic, the apoplast being the main site of their accumulation (van Loon, 1999). Apart from being present in the primary and secondary cell walls of infected plants, PRs are also found in cell wall appositions (papillae) deposited at the inner side or the space of cell wall in response to fungal attack. It was relatively conserved in evolution, the same type of PR proteins of different plants were highly similar in the molecular structure, amino acid composition and so on. So the E-value of homologous sequences less than e-100 was seemed as the gene copies. Table 5 showed the gene mapping, the number of sequence copy, the analysis number and length of gene, and their number of exon.
 Table 4 The information of the members in PR family |
We studied all the members of PR protein family and linkage group distribution of their copies, and found that PR protein mainly distributed on Gm05, Gm10, Gm13, Gm15, Gm17, Gm19, and Gm20 linkage groups, while less on others. It demonstrated that PR protein genes were clustered distribution on the linkage groups, especially among the members in the same family, as shown in Figure 3, the most members of PR5 family located in Gm10 linkage group (Figure 2; Figure 3).
.png) Figure 2 Distribution of genes in PRs families on the LGs |
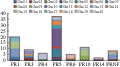 Figure 3 Distribution of PRs families on the LGs |
The phylogenetic analyses for thirty-six members in 9 families by using MEGA4 showed that most of the members in the same family had the similar evolutionary origin, such as the PR4 family was all clustered together and each of them was very short in distance, the distance between PR4-2 and PR4-4 was less than 0.002; moreover, there was also a far evolutionary relationships between the members in the same family, such as PR1-4 and PR1-3. However, the PRNF may have evolved from some other families (Figure 4).
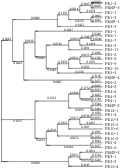 Figure 4 The cluster analysis of PR protein gene |
2 Discussion
2.1The significance and feasibility of PR protein gene prediction
The expression of PR proteins was regulated by pathogen infection, plant development, and other factors such as stress or hormones, which involved in partial and systemic resistance. However, it is seldom known about the regulation mechanism of gene expression and the gene expression in the pathway of signal transduction. Therefore, HMMER has become the powerful method based on sequence alignment and characteristic analysis. Meantime, the PR protein sequences were conserved and the function of the same family was similar. The combination of HMMER and BLAST produced a lot of repeat sequences which verified the accuracy of the two methods. However, both methods present the phenomenon of less result after prediction; there might be three kinds of reasons as follows. Firstly, the higher homology among the PR protein family members, the less it makes the source for researching; secondly, because of the higher homology, the low-scoring sequence was excluded; as well as the principle of HMMER sequence prediction was also the common features integration among different sequences. And the feature was also used for sequence searching, which also reduced the source of sequence.
2.2 PR protein characterization analysis
The basic mechanism of PR protein gene expression is transcriptional activation. Most members of the same family and the different copies have the same number of intron. Different introns were different in length, which decided whether to accept some kind of signaling. Most members are clustered distribution on several linkage groups, which may induce amounts of protein production continuously after pathogen infection or may stimulate the expression after receiving a strong signal. Due to the homology in different families, they can receive the same stimulus signal to activate the expression and the repeat sequences in different families may also be deduced after prediction. The activation of expression of different families stimulated by the same signal not only synchronization and coordination, but also mutual inhibition. The target sequence performed specificity on the signal, for example, there are three kinds of tomato PR proteins, which response to the three isomers of γ-aminobutyric acid (GABA) differences reaching about 86%, indicating that the differences between the different families may determine the sequence specificity of PR proteins (Zhao and Guo, 2003).
3 Materials and Methods
3.1 Materials
3.1.1 The collection and collation of PR protein sequences
The Arabidopsis (Arabidopsis thaliana), maize (Zea mays), rice (Oryza sativa), and legumes (Fabaceae) PR protein sequences were downloaded from the NCBI (http://www.ncbi.nlm.nih.gov/) and classified according to their families.
In this document, the number of PR protein sequences for Arabidopsis, rice, corn, and legumes downloaded from the internet were 114, 83, 23, and 46, respectively. Among them, only 4 PR proteins belonged to soybean. All of them were classified by their family names, and we totally obtained 266 members belonged to 9 families, which were 1, 2, 3, 4, 5, 6, 10, 14 and uncertain family (Pathogenesis-related protein in No Family, PRNF) (Table 5).
 Table 5 Classification and numbers of PR families Note: PRNF means pathogenisis related protein in no family |
3.1.2 Database of soybean protein and software preparation
Soybean genome database was downloaded from NCBI (http://www.ncbi.nlm.nih.gov/), the open reading frame (ORF) in the genome were predicted by GENSCAN and then were translated into protein to establish the protein database. The soybean EST database was downloaded at the same time.
Download the blast 2.2.16 package for the local alignment from NCBI, and HMMER 3.0 software for installment.
3.2 Methods
3.2.1 PR protein were predicted by BLAST
The repeat sequences of the PR protein would be removed. The sequence can be identified as the homology sequence when their E-value was less than 0.01, and only a non-repetitive sequence was retained in accordance with the soybean genetic relationship options from near to far. The unique PR protein sequence would be BLAST with the database of soybean EST by using tBLASTn procedure, and the result sequences from soybean EST that homology with the PR protein were determined to be the candidates of PR proteins.
3.2.2 HMMER predicted PR protein
The multiple alignment according to the PR family were made among the download sequences of Arabidopsis, maize, rice, and legumes to form the ALIGN file and converted it into a recognizable HMMER file. They were saved as seed and align files respectively. For the families with less members or less homologous sequences after alignment, we would search through NCBI to identify the other homology genes in the network database, and then carried out the multiple alignment to form the seed file.
The file of align and seed were transformed into hidden Markov model (HMM) file seed.hmm and align.hmm by HMMbuild, and established the HMM of the family-owned of PR proteins.
The program commanded as "# hmmbuild PR.hmm_PR.msf".
The HMM files were compared with the database of soybean protein by HMMsearch, according to the default E-value 0.01, and obtained the out file.
The program commanded as "# hmmsearch PR.hmm soybeandatabase> PR.out".
According to the out file, the predicted peptide sequences and CDS sequences of PR protein can be found in the new local protein database, as the candidate of soybean PR protein.
3.2.3 Prediction and analysis of PR protein sequence of soybean
The candidate soybean EST sequences of PR protein predicted through two ways were integrated, and the repeat sequences were removed. The candidate sequences were also assembled for several rounds and extended by GENSCAN (http://genes.mit.edu/GENSCAN.html) to predict the full-length ORF. The predicted full-length ORF were annotated by researching with the NCBI to determine the function and the real CDS. At last they were classified according to their family or order and named the soybean PR protein.
The PR protein of soybean were mapped on soybean genome by Phytozome (http://www.phytozome.net/), and determined their distribution on the genome, number of copies, the number of exon and intron, and structural and evolution variation among more copies of genes.
Author's contributions
Qingshan Chen professor was responsible for experimental design and the experiment direction; Jing Wang and Liwei Zhang were responsible for software analysis, data management and paper writing; Chunyan Liu, Yuhua Li and Guohua Hu teachers helped modify the paper.
Acknowledgements
This study was funded by the National Natural Science Foundation (30971809), and we got the help and support from Zhu Mingxi, Thanks a lot.
References
Batalia M.A., Monzingo A.F., Roberts W., and Robertus J.D., 1996, The crystal structure of the antifungal protein zeamatin,a member of the thaumatin-like, PR-5 protein family, Nature Struct. Biol., 3: 19-23
http://dx.doi.org/10.1038/nsb0196-19
PMid:8548448
Bol J.F., Linthorst H.J.M., and Cornelissen B.J. C., 1990, Plant pathogenesis—related proteins induced by virus infection, Annu. Rev. Phytopathol., 28: 113-138
http://dx.doi.org/10.1146/annurev.py.28.090190.000553
http://dx.doi.org/10.1146/annurev.phyto.28.1.113
Edreva A., 2005, Pathogenesis-related proteins: research progress in the last 15 years, Gen. Appl. Plant Physiology, 31(1-2): 105-124
Graham M.Y., 2005, The diphenylether herbicide lactofen induces cell death and expression of defense-related genes in soybean, Plant Physiol., 139(4): 1784-1794
http://dx.doi.org/10.1104/pp.105.068676
PMid:16299178 PMCid:1310559
van Loon L.C., and van K., 1970, Polyacrylamide disc electrophoresis of the soluble leaf proteins from Nicotiana tabacum var. Samsun and Samsun NN. II. Changes in protein constitution after infection with tobacco mosaic virus, Virology, 40: 199-211
http://dx.doi.org/10.1016/0042-6822(70)90395-8
van Loon L.C., and van Strien E.A., 1999, The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins, Physiol. Mol. Plant Pathol., 55: 85-97
http://dx.doi.org/10.1006/pmpp.1999.0213
van Loon L.C., Pierpoint W.S. and Boller T., 1994, Recommendations for naming plant pathogenesis-related proteins, Plant Mol. Biol. Rep., 12(3): 245-264
http://dx.doi.org/10.1007/BF02668748
Wang J., 1995, Recent advance of plant disease resistance. Zhiwu Shenglixue Tongxun (Plant Physiology Communications), 31 (4): 312-317
Wen Y.J., Huang Q.S., Liang S., Bin J.H., and He H.W., 2008, Roles of pathogenesis-relative protein 10 in plant defense response, Zhiwu Shenglixue Tongxun (Plant Physiology Communications),44(3): 585-592
Zhao S.Q., and Guo J.B., 2003, Systemic acquired resistance and signal transduction in plant, Zhongguo Nongye Kexue (Science Agriculture Sinica), 36(7): 781-787
. PDF(430KB)
. HTML
Associated material
. Readers' comments
Other articles by authors
. Jing Wang
. Liwei Zhang
. Chunyan Liu
. Yuhua Li
. Qingshan Chen
. Guohua Hu
Related articles
. Soybean ( Glycine max L.)
. Pathogenesis related proteins (PRs)
. BLAST
. HMMER
Tools
. Email to a friend
. Post a comment


