The Neurospora crassa cmd, trm-9, and nca-2 Genes Play a Role in Growth, Development, and Survival in Stress conditions 
 Author
Author  Correspondence author
Correspondence author
Genomics and Applied Biology, 2015, Vol. 6, No. 7 doi: 10.5376/gab.2015.06.0007
Received: 24 Apr., 2015 Accepted: 02 Aug., 2015 Published: 14 Oct., 2015
Laxmi V. and Tamuli R., 2015, The Neurospora crassa cmd, trm-9, and nca-2 Play a Role in Growth, Development, and Survival in Stress conditions, Genomics and Applied Biology, Vol.6, No.7, 1-8 (doi: 10.5376/gab.2015.06.0007)
The calmodulin protein antagonists, trifluoperazine (TFP) and chlorpromazine (CPZ) inhibit the growth, carotenoids accumulation and sexual development of Neurospora crassa. In addition, N. crassa strains lacking trm-9, a cation-ATPase, showed defect in growth. Moreover, strains lacking both trm-9 and another Ca2+-ATPase nca-2, exhibited a severe growth defect, an increased sensitivity to CaCl2, and a reduction in acquisition of thermotolerance induced by heat shock temperature. Therefore, the cmd, trm-9, and nca-2 play a role in growth, survival in calcium stress and induced heat shock temperature in N. crassa.
Introduction
Calcium (Ca2+) signaling is involved in regulating numerous processes in eukaryotes ranging from fungi to mammals. The Ca2+-signaling process is initiated primarily due to transient raise in concentration of cytosolic free Ca2+ ([Ca2+] c), which is recognized by Ca2+ sensor proteins. One of the versatile and evolutionary conserved Ca2+-sensor proteins is calmodulin (CaM), which binds Ca2+ with high affinity and specificity. CaM plays an important role in modulating DNA repair, DNA synthesis, cell proliferation, cyclic nucleotide and glycogen metabolism, secretion, motility and Ca2+ transport (Means and Dedman 1980; Smallwood et al., 2009). CaM also plays an important role for the regulation of stress response pathways in pathogenic fungi Candida albicans and Cryptococcus neoformans (Kraus and Heitman, 2003). In the budding yeast Saccharomyces cerevisiae, CaM is required for mitotic progression and acquisition of induced thermotolerance (Iida et al., 1995). Similar to the S. cerevisiae, in the filamentous fungi Aspergillus nidulans, CaM is critical for the progression through the G2/M transition (Kahl and Means 2003).
The filamentous fungus Neurospora crassa has a unique calcium (Ca2+) signaling machinery, CaM is encoded by NCU04120 that appears to be an essential gene for viability (Galagan et al., 2003; Borkovich et al., 2004; Tamuli et al., 2013). In N. crassa, unlike the vertebrate counterparts, only one CaM gene has been identified (Capelli et al., 1993; Cox et al., 1982; Perez et al., 1981; Galagan et al., 2003). In vertebrates, CaM protein is encoded by multiple genes, for example, six genes have been detected in zebra fish, three genes in human and rat, two genes in frog and two genes in chicken (Luan et al., 2007). Coding sequence of the CaM encoding gene NCU04120 contains six exons and five introns, and CaM possesses conserved EF-hand domains (Tamuli et al., 2013). In N. crassa CaM antagonists, trifluoperazine (TFP) and chlorpromazine (CPZ) caused shortening of period length of the conidiation rhythm and light induced phase shifting (Sadakane and Nakashima 1996; Suzuki et al., 1996). In addition, possible role of CaM in activation of chitin synthase enzyme in N. crassa was studied by examining the effects of TFP on protoplast regeneration (Suresh and Subramanyam, 1997).
One of the targets of CaM is the Ca2+ -ATPase, a Ca2+ pump that help in fine tuning of Ca2+ homeostasis in cells by pumping Ca2+ out of cells. Ca2+ -ATPases hydrolyze ATP to catalyze active Ca2+-efflux across biological membranes, and maintain a steep Ca2+ gradient across the plasma membrane (Hao et al., 1994). CaM stimulates plasma membrane Ca2+-ATPase (PMCA) activity by binding to an autoinhibitory domain of PMCA. The CaM-binding domain is located near the C-terminus of PMCA (Osborn et al., 2004; Giacomello et al., 2013). Besides interacting with (Ca2++Mg2+) -activated ATPase in isolated cardiac sarcoplasmic reticulum and RBC membrane, CaM also interacts with ciliary dynein ATPase of Tetrahymena (Blum et al., 1980; Kirchberger and Antonetz, 1982; Lopes et al., 1990). In plants, Ca2+-activated CaM regulates different Ca2+-ATPases (Peerseen et al., 1997; Harper et al., 1998; Hong et al., 1999; Chung et al., 2000; Malmström et al., 1997, 2000). In N. crassa, nine ATPases have been identified and they possess conserved cation transporter/ATPase domain in the proteins (Galagan et al., 2003; Borkovich et al., 2004; Tamuli et al., 2013). N. crassa ATPases are found distributed in different branches during a phylogenetic analysis, NCA1 in ERCA, NCA2 and NCA3 in PMCA, PMR1 in PMR1, and PH-7 in ENA branch (Haro et al., 1991; Benito et al., 2000). Lack of NCA-2 results in slow growth, Ca2+ sensitivity, female sterility, and accumulation of more Ca2+ than the wild-type; indicating that it functions in the plasma membrane to pump Ca2+ out of the cell (Bowman et al., 2011). NCA-2 is more similar to the PMC-type proteins of animal cell than the Pmc1p in S. cerevisiae that resides in the vacuole (Bowman et al., 2011). In addition, one of the cation-ATPases trm-9, which is encoded by the gene NCU04898, shows sequence homology to spf1 gene of S. cerevisiae. SPF1 family ATPases genes are conserved from yeast to human; however, the functions of these ATPases remain unclear. SPF1 is not essential for cell viability and its substrate specificity is unknown and loss of SPF1 may perturb homeostasis of ions that affects modification and sorting of proteins in the secretory pathway of yeast (Cronin et al., 2000; Suzuki, 2001).
To investigate the cellular role of CaM in N. crassa, we used CaM antagonists, TFP and CPZ. Moreover, we studied two other genes trm-9 and nca-2 using their knockout mutants. We found that the cmd, trm-9, and nca-2 genes play a role in growth, Ca2+ sensitivity, and in acquisition of thermotolerance induced by heat shock temperature in N. crassa.
Materials and Methods
Sequence analysis
BLAST (Altschul et al., 1990) analysis was performed using software tools available from NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and the proteins were selected based on E value, % identities and gapes as described previously (Tamuli et al., 2011). The Conserved Domain Database (CCD) (Marchler- Bauer et al., 2009) database was used to identify conserved domains in the protein. The homologue protein sequences were aligned with ClustalX 1.83 (Thompson et al., 1997) and visualized using GeneDoc (Nicholas et al., 1997). Phylogenetic trees were constructed from the aligned sequences using the minimum-evolution method (Rzhetsky and Nei, 1992), 500 bootstrap replications as test of phylogeny (Felsenstein 1985) and the software MEGA5 (Tamura et al., 2011). Promoter region was analyzed by selecting ~2 kb sequences from upstream of Transcription Start Site and transferred to MatInspector in Genomatix software (http://www.genomatix. de/cgi- bin//matinspector_prof/mat_fam) to predict transcription factor binding sites (Quandt et al., 1995).
Strains, growth, crosses maintenance
N. crassa wild-type strains 74-OR23-1 mat A (FGSC 987), 74-OR8-1 mat a (FGSC 988), Ca2+ signaling mutant strain ∆NCU04898.2::hph mat A (FGSC 1304 0), and ∆NCU04736.2::hph mat A (FGSC 13071) were generated by the Neurospora genome project and obtained from the Fungal Genetics Stock Center (FGSC; University of Missouri, Kansas city, MO 64110) (Colot et al., 2006; McCluskey 2010). The ∆NCU04898.2::hph ∆NCU04736.2::hph double mutant was generated by crossing the individual single mutant strain, and presence of the knockout alleles were verified using polymerase chain reactions (PCR) of the progeny strains (Supplementary Figure 1).
Growth, crossing, and maintenance of Neurospora strains were essentially as described by Davis and De Serres (1970). The apical growth was analyzed by using standard race tube assay and calculated as cm h-1 (Ryan et al., 1943, 1950). For aerial hyphae, ~1 X 106 cells/ml of each strain was grown in liquid Vogel’s sucrose media (VSM) and incubated at 30C for 48 h in dark followed by 24 h light illumination at room temperature and height of aerial hyphae was measured (Deka and Tamuli, 2013). Conidial count was done after 72 hours of growth; a sample of each strain was withdrawn and harvested using sterile water followed by conidial counting using a haemocytometer under a Trinocular Phase Contrast Microscope (Supplementary Figure 2). For growth yield, ~1 X 106 cells/ml of each strain were inoculated in liquid Vogel’s medium at 30ºC with shaking at 200 rpm for growth. Mycelia were collected at a regular interval of 24 h by filtration, dried and weighed over a period 96 h. For analysis of hyphal morphology, strains were grown for 12 h on a thin layer of Vogel’s agar on glass slide, and observed under microscope at 20X magnification. In addition, statistical significance was performed according to variance analysis (ANOVA, P < 0.05).
Assay for calcium sensitivity and thermotolerance
Assay for calcium sensitivity was done essentially as described previously (Deka et al., 2011). Briefly, conidia was placed in the centre of petri dishes containing Vogel’s glucose (1.5%) media supplemented with 0.0 M, 0.2 M, 0.3 M, 0.4 M CaCl2 incubated at 30ºC and colony diameter was measured every 3 h over a period of 24 h and growth rates were calculated as cm h-1. For measuring thermotolerance, three days-old conidia were inoculated into liquid Vogel’s Medium at a concentration of ~1 X 106 cells/ml and germinated for 2 h with shaking at 200 rpm at 30C. These germlings were exposed to different heat treatment condition in two sets one set was held at 30C for uninduced condition and the other set at 44C for induced condition for 30 min, then one set of each were given a lethal heat shock at 52C for 20 min. (Yang Qi and Borkovich, 1999; Kumar and Tamuli, 2014). After that these conidia were spread on sorbose agar (0.05 % fructose, 0.05 % glucose, 2% sorbose, 2 % Bacto agar) plate and incubated at 30C for 2 days. Percent survival was obtained by dividing the number of viable colonies on plates subjected to heat treatment by the number of viable colonies on plates held at 30C (control) and multiplying by 100.
Carotenoid accumulation
To measure carotenoid accumulation, ~1 X 106 cells/ml of each N. crassa strains were inoculated into Vogel’s sucrose (2%) medium supplemented with 0.2% Tween 80 used as a wetting agent to prevent conidiation (Zalokar, 1954) and kept for growth at 30C for two days in dark, and at room temperature for one day under light. After that mycelia were filtered, lyophilized and powdered. Carotenoids were extracted from 50 mg lyophilized powder by using acetone and hexane. Total carotenoids content were calculated by measuring the maximum absorbance value at 470 nm and using formula: Total carotenoid content (µg/g) = [Total absorbance x Total volume of extract (ml) x 104]/ [Absorbance coefficient (2500) x Sample weight (g)] (Rodriguez-Amaya and Kimura 2004).
Results and Discussion
NCU04120, NCU04898, and NCU04736 genes encode CaM, TRM-9, and NCA-2, respectively, and contain conserved domains
The N. crassa calmodulin gene (cmd) NCU04120 encodes a highly conserved Ca2+ signaling protein CaM that possesses four conserved EF hand motifs (Supplementary Figure 3A). Similarly, the NCU04898 encodes a Ca2+/cation ATPases annotated as TRM-9 (http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html). The TRM-9 possesses one conserved E1-E2 ATPases domain, and one halo-acid dehalogenase like hydrolase domain (Supplementary Figure 3B). In addition, phylogenetic analysis revealed that CaM and TRM-9 proteins are clustered with homologues from related (Supplementary Figure 4). Promoter analysis revealed important putative regulatory elements involved in transcription of cmd and trm-9 gene by using MatInspector software (Supplementary Figure 5). The NCU04736 gene was previously shown to encode nca-2 (Bowman et al., 2009; Bowman et al., 2011).
The cmd, trm-9, and nca-2 genes are involved in growth
We used CaM antagonists, trifluoperazine (TFP) and chlorpromazine (CPZ) to study the effect of CaM inhibition on growth of wild-type strain of N. crassa. We found that both TFP and CPZ inhibit growth, a hyphal branching, and development of aerial hyphae in N. crassa (Figure 1). Moreover, addition of TFP (10, 20, 40, 60, 80, and 100 µM) or CPZ (20, 40, 60, 80, 100 µM) in the synthetic crossing medium (SCM) causes a defect in perithecia formation in N. crassa and results in a sterile phenotype (data not shown). In addition, the cmd transcript level was found to be decreased in the presence of TFP and CPZ as revealed by the real time PCR analysis (Supplementary Figure 6). These results suggest that CaM play a role in vegetative growth, hyphal development, and sexual development in N. crassa.
.png) Figure 1 Effect of trifluoperazine (TFP) and chlorpromazine (CPZ) on growth of N. crassa. (A) Effect of TFP and CPZ at various concentrations on apical growth. (B) Abnormal hyphal morphology with increasing concentrations of TFP and CPZ. (C) Aerial hyphae length of cultures grown for 72 h in various concentrations of TFP and CPZ. Error bars indicate the standard errors calculated from the data for three independent experiments. Statistically significant values are indicated by asterisks, *P < 0.05 |
The △trm-9 mutant displayed a slow growth phenotype (Figure 2A). However, the slow growth phenotype of the △trm-9 mutant was not due to a defect in the ergosterol profile (Supplementary Figure 7) and the growth of the △trm-9 mutant was not affected by addition of various amounts of CaCl2, sucrose and NaCl in the medium indicating that the △trm-9 mutant is insensitive to these stress conditions (data not shown). The growth defect was more severe in the △trm-9 △nca-2 double mutant that showed distinct colony morphology with matty-like colony growth (Figure 2B). In addition, the dry weight of the strains followed the order wild-type>△nca-2> △trm-9> △trm-9 △nca-2 (Figure 2C). The △trm-9 △nca-2 double mutant also showed sensitivity to CaCl2, reduced aerial hyphae development and ultraviolet (UV) survival (Supplementary Figure 8). Therefore, these results suggested that lack of both trm-9 and nca-2 result in impaired growth, hyphae development and conidial development in N. crassa.
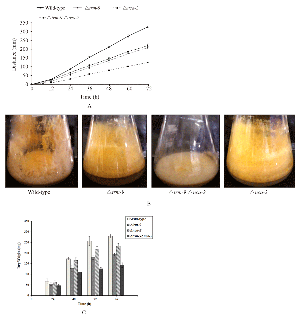 Figure 2 Growth phenotypes. (A) Rate of apical growth of the wild-type, △trm-9, △nca-2, and △trm-9nca-2 strains were measured using race tubes. Growth rate of △trm-9 △nca-2 double mutant strain was lesser as compared to parental single mutants and wild-type strain. (B) Colony morphology of wild-type, △trm-9, △nca-2, and △trm-9nca-2 double mutant strains. The △trm-9△nca-2 double mutant strain showed matty-like growth and reduced pigmentation. (C) Dry weight of △trm-9, △nca-2, △trm-9 △nca-2 and wild-type strains. Dry weight yield of △trm-9 △nca-2 double mutant strain was less than parental single mutants and the wild-type |
Carotenoids accumulation
We also analyzed carotenoid accumulation in N. crassa in the presence of TFP and CPZ to investigate the role of CaM in carotenoid accumulation. The carotenoid profile of the wild-type strain in presence of the inhibitors followed the order wild-type > 10 µM CPZ > 20 µM CPZ >10 µM TFP > 20 µM TFP (Figure 3A). Therefore, these results indicate the CaM protein might modulate carotenoid accumulation in N. crassa. The difference of carotenoids accumulation in presence of TFP and CPZ might be due to the difference of mechanism of inhibition mediated by TFP and CPZ. Furthermore, carotenoids accumulation in the △trm-9 △nca-2 double mutant was lower than either of the parental single mutant strains, and reduced further on medium supplemented with high concentrations of CaCl2 (Figure 3B). In addition, △nca-2 mutant was unable to grow on medium supplemented with 0.3 M CaCl2 or more (supplementray Figure 8A) and consequently, no carotenoids was accumulated; however, accumulation of carotenoids in the △trm-9 was similar to the wild-type (Figure 3B). Therefore, these results suggest that nca-2 plays a role in carotenoids biosynthesis.
|
Figure 3 Analysis of carotenoids content. (A) Carotenoid content of wild-type strain in the presence of CaM antagonist TFP and CPZ. Standard errors calculated from the data for three independent experiments are shown using error bars. TFP act as negative regulator for carotenoid accumulation whereas CPZ act as positive regulator during carotenoid accumulation. (B) Carotenoids accumulation of wild-type, △trm-9, △nca-2, and △trm-9 △nca-2 double mutant strains during Ca2+stress. Carotenoids extracted from these strains grown in Vogel’s liquid medium without CaCl2, supplemented with various concentrations of CaCl2. Carotenoids were extracted and estimated in µg carotenoids per g of dry weight. Error bars show the standard errors calculated from the data for three independent experiments. Statistically significant values are indicated by asterisks, *P < 0.05 |
Lack of both nca-2 and trm-9 affect in acquisition of induced thermotolerance
We studied the ability of the △trm-9 △nca-2 double mutant in acquisition of induced thermotolerance. The △trm-9 △nca-2 double mutants showed decreased survival in induced thermotolerance as compared to parental single mutants. The survival in induced heat shock temperature followed the order △nca-2> △trm-9>△trm-9 △nca-2>wild-type (Figure 4). Therefore, lack of both nca-2 and trm-9 had a negative effect in acquisition of induced thermotolerance.
 Figure 4 Thermotolerance measurement of wild-type, △trm-9, △nca-2, and △trm-9 △nca-2 double mutant strains in induced (44°C) and uninduced (30°C) conditions. Each data point represents the mean of three independent experiments |
Conclusions
CaM and its target proteins mediate diverse cellular functions. The CaM antagonists TFP and CPZ affect growth, aerial hyphae development, carotenoids accumulation and sexual development in N. crassa. In addition, △trm-9 mutant has a slow growth phenotype and less dry weight content. Moreover, the △trm- 9 △nca-2 double mutant showed a severe growth defect, less carotenoid accumulation, reduced conidial count, an increased sensitivity to CaCl2, and reduced viability in acquisition of thermotolerance induced by heat shock temperature. Thus, in this study, we have shown that cmd, trm-9, and nca-2 genes play an important role in growth, pigmentation, and stress- tolerance in N. crassa.
Acknowledgements
The Fungal Genetic Stock Center (FGSC) generously waived charges for strains and race tube. The FGSC was supported by National Science Foundation grant BIR-9222772. We thank Upasana Sarma for performing some initial experiments. VL was supported by a Research Fellowship from the Ministry of Human Resource Development, Government of India. This work was supported partially by grants, including BT/PR3635/ BCE/8/ 892/2012, from the Department of Biotechnology, Government of India.
References
Altschul S.F., Gish W., Miller W., Myers E.W., et al., 1990, Basic local alignment search tool, J Mol Biol, 215:403-410
http://dx.doi.org/10.1016/S0022-2836(05)80360-2
Benito B., Garciadeblas B., et al., 2000, Molecular cloning of the calcium and sodium ATPases in Neurospora crassa, Mol. Microbiol, 35:1079-1088
http://dx.doi.org/10.1046/j.1365-2958.2000.01776.x
Blum J.J., Hayes A., Jamieson G.A. Jr., Vanaman T.C., 1980, Calmodulin confers calcium sensitivity on ciliary dynein ATPase, J Cell Bio, 87:386-397
http://dx.doi.org/10.1083/jcb.87.2.386
Borkovich K.A., Alex L.A., Yarden O., Freitag M., et al., 2004, Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism, Microbiol Mol Biol Rev, 68:1-108
http://dx.doi.org/10.1128/MMBR.68.1.1-108.2004
Bowman B.J., Draskovic M., Freitag M., Bowman E.J., 2009, Structure and distribution of organelles and cellular location of calcium transporters in Neurospora crassa, Eukaryot Cell, 8:1845-1855
http://dx.doi.org/10.1128/EC.00174-09
Bowman B.J., Abreu S., Margolles-Clark E., Draskovic M., et al., 2011, Role of four calcium transport proteins, encoded by nca-1, nca-2, nca-3, and cax, in maintaining intracellular calcium levels in Neurospora crassa, Eukaryot Cell, 10: 654-661
http://dx.doi.org/10.1128/EC.00239-10
Capelli N., Tuinen V.D., Perez O.R., Arrighi J.F., Turian G., 1993, Molecular cloning of a cDNA encoding CaM from Neurospora crassa, FEBS 321:63-68
http://dx.doi.org/10.1016/0014-5793(93)80622-2
Chung W.S., Lee S.H., Kim J.C., Heo W.D., Kim M.C., Park C.Y., Park H.C., Lim C.O., Kim W.B., Harper J.F., Cho M.J., 2000, Identification of a calmodulin-regulated soybean Ca2+ -ATPase (SCA1) that is located in the plasma membrane, Plant Cell, 12:1393-1407
http://dx.doi.org/10.1105/tpc.12.8.1393
Colot H.V., Park G., Turner G.E., Ringelberg C., Crew C.M., Litvinkova L., Weiss R.L., Borkovich K.A., Dunlap J.C., 2006, A high throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors, ProcNatlAcadSci USA 103:10352–10357
http://dx.doi.org/10.1073/pnas.0601456103
Cronin S.R., Khoury A., Ferry D.K., Hampton R.Y., 2000, Regulation of HMG-CoA reductase degradation requires the P-Type ATPase Cod1p/Spf1p, J Cell Bio 148: 915-924
http://dx.doi.org/10.1083/jcb.148.5.915
Cox J.A., Ferraz C., Demaille J.G., Perez R.O., Van Tuinen D., Marmé D., 1982, Calmodulin from Neurosporacrassa. General properties and conformational changes, J Biol Chem, 257:10694-10700
Davis R.H., De Serres F.J., 1970, Genetic and microbial research techniques for Neurospora crassa, Methods Enzymol, 17:79-143
http://dx.doi.org/10.1016/0076-6879(71)17168-6
Deka R., Kumar R., Tamuli R., 2011, Neurospora crassa homologue of neuronal calcium sensor-1 has a role in growth, calcium stress tolerance, and ultraviolet survival, Genetica 139:885-894
http://dx.doi.org/10.1007/s10709-011-9592-y
Deka R., Tamuli R., 2013, Neurospora crassa ncs-1, mid-1 and nca-2 double-mutantphenotypes suggest diverse interaction among three Ca2+-regulating gene products, J Genet, 92:559-563
http://dx.doi.org/10.1007/s12041-013-0270-y
Felsenstein J., 1985, Confidence limits on phylogenies: an approach using
the bootstrap, Evolution, 39:783-791
http://dx.doi.org/10.2307/2408678
Galagan J.E., Calvo S.E., Borkovich K.A., Selker E.U., et al., 2003, The genome sequence ofthe filamentous fungus Neurospora crassa, Nature 422:859–868
http://dx.doi.org/10.1038/nature01554
Giacomello M., De Mario A., Scarlatti C., Primerano S., Carafoli E., 2013, Plasma membrane calcium ATPases and related disorders, J Biochem and Cell Bio 45:753-762
http://dx.doi.org/10.1016/j.biocel.2012.09.016
Hao L., Rigaud J.L., Inesi G., 1994, Ca2+/H+countertransport and electrogenicity in proteoliposomes containing erythrocyte plasma membrane Ca- ATPase and exogenous lipids, J Bio Chem, 269:14268-14275
Haro R., Garciadeblas B., Rodríguez-Navarro A., 1991, A novel P-type ATPase from yeast involved in sodium transport, FEBS Lett, 291: 189-191
http://dx.doi.org/10.1016/0014-5793(91)81280-L
Harper J.F., Hong B., Hwang I., Guo H.Q., Stoddard R., Huang J.F., Palmgren M.G., Sze H., 1998, A novel calmodulin-regulated Ca2+-ATPase (ACA2) from Arabidopsis with an N-terminal autoinhibitory domain, J Biol Chem, 273:1099-10106
http://dx.doi.org/10.1074/jbc.273.2.1099
Hong B., Ichida A., Wang Y., Gens J.S., Pickard B.G., Harper J.F., 1999, Identification of a calmodulin-regulated Ca2+-ATPase in the endoplasmic reticulum, Plant Physio, 119:1165-1176
http://dx.doi.org/10.1104/pp.119.4.1165
Iida H., Ohya Y., Anraku Y., 1995, Calmodulin-dependent protein kinase II and calmodulin are required for induced thermotolerance in Saccharomyces cerevisiae, Curr Genet, 27:190-193
http://dx.doi.org/10.1007/BF00313434
Kahl C.R., Means A.R., 2003, Regulation of cell cycle progression by calcium/CaM-dependent pathways, Endocrine society, 24:719-736
Kirchberger A.M., Antonetz T., 1982, Calmodulin-mediated regulation of calcium transport and (Ca2+ + M2+)-activated ATPase activity in isolated cardiac sarcoplasmic reticulum, J Bio Chem, 257: 5685-5691
Kraus R.P., Heitman J., 2003, Coping with stress: CaM and calcineurin in model and pathogenic fungi, Biochem Biophy Res, 311:1151-1157
http://dx.doi.org/10.1016/S0006-291X(03)01528-6
Kumar R., Tamuli R., 2014, Calcium/calmodulin-dependent kinases are involved in growth, thermotolerance, oxidative stress survival, and fertility in Neurospora crassa, Archives of Microbio, 196: 295-305
http://dx.doi.org/10.1007/s00203-014-0966-2
Lopes M.C., Vale M.G., Carvalho A.P., 1990, Ca2+ -dependent binding of tamoxifen to calmodulin isolated from bovine brain, Cancer Res, 50:2753-2758
Luan J., Liu Z., Zhang S., Li H., Fan C., Li L., 2007, Characterization, evolution and expression of the calmodulin1 genes from the amphioxus Branchiostomabelcheritsingtauense, Acta Biochem Biophys Sin (Shanghai). 39:255-264
http://dx.doi.org/10.1111/j.1745-7270.2007.00277.x
Malmstrom S., Askerlund P., Palmgren M.G., 1997, A calmodulin-stimulated Ca2+ -ATPase from plant vacuolar membranes with a putative regulatory domain at its N-terminus, FEBS Letters, 400:324-328
http://dx.doi.org/10.1016/S0014-5793(96)01448-2
Malmstrom S., Akerlund H.E., Askerlund P., 2000, Regulatory role of the N terminus of the vacuolar calcium-ATPase in cauliflower, Plant Physiol, 122: 517-526
http://dx.doi.org/10.1104/pp.122.2.517
Marchler-Bauer A., Anderson J.B., Chitsaz F., Derbyshire M.K., et al., 2009, CDD: specific functional annotation with the Conserved Domain Database, Nucleic Acids Res 37 (Database issue):D205-D210
http://dx.doi.org/10.1093/nar/gkn845
McCluskey K., Wiest A., Plamann M., 2010, The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research, J Biosci, 35:119-126
http://dx.doi.org/10.1007/s12038-010-0014-6
Means R.A., Dedman R.J., 1980, CaM-an intracellular calcium receptor, Nature, 285:73-77
http://dx.doi.org/10.1038/285073a0
Nicholas K.B., Nicholas H.B., 1997, GeneDoc: a tool for editing and annotating multiple sequence alignments, Distributed by the author, http://www.psc.edu/biomed/genedoc
Osborn D.K., Zaidi A., Mandal A., Urbauer R.J., Johnson C.K., 2004, Single molecule dynamics of the calcium-dependent activation of plasma membrane Ca2+-ATPase by calmodulin, Biophy J., 87: 1892-1899
http://dx.doi.org/10.1529/biophysj.103.039404
Peersen B.O., Madsen S.T., Falke J.J., 1997, Intermolecular tuning of calmodulin by target peptides and proteins: Differential effects on Ca2+ binding and implications for kinase activation, Protein Science, 6:794-807
http://dx.doi.org/10.1002/pro.5560060406
Perez O.R., Tuninen V.D., Marme D., Cox A.J., Turian G., 1981, Purification and identification of CaM from Neurospora crassa, FEBS 133:205-208
http://dx.doi.org/10.1016/0014-5793(81)80506-6
Quandt K., Frech K., Karas H., Wingender E., et al., 1995, MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data, Nucleic Acids Res, 23:4878-4884
http://dx.doi.org/10.1093/nar/23.23.4878
Rodriguez-Amaya D.B., Kimura M., 2004, Harvest Plus handbook for carotenoid analysis. Harvest Plus Technical Monograph 2, International Food Policy Research Institute (IFPRI) and International Center for Tropical Agriculture (CIAT), Washington, DC
Ryan F.J., Beadle G.W., Tatum E.L., 1943, the tube method of measuring the growth rate of Neurospora, Am J Bot, 30:784-799
http://dx.doi.org/10.2307/2437554
Ryan F.J., 1950, Selected method of Neurospora genetics, Method Med Res, 3:51-75
Rzhetsky A., Nei M., 1992, Statistical properties of the ordinary least-generalized least-squares, and minimum-evolution methods of phylogenetic inference, J Mol Evol, 35:367-375
http://dx.doi.org/10.1007/BF00161174
Sadakane Y., Nakashima H., 1996, Light-induced phase shifting of the circadian conidiation rhythm is inhibited by calmodulin antagonists in Neurospora crassa, J Biol Rhythms, 11:234-240
http://dx.doi.org/10.1177/074873049601100305
Smallwood S.H., Lopez F.D., Eberlein E.P., Watson J.D., and Squier C.T., 2009, Calmodulin mediates DNA repair pathways involving H2AX in response to low-dose radiation exposure of RAW 264.7 Macrophages, Chem Res Toxicol, 22:460-470
http://dx.doi.org/10.1021/tx800236r
Suresh K., Subramanyam C., 1997, A putative role for CaM in the activation of Neurospora crassa chitin synthase, FEMS Micro biol letters , 150:95-100
http://dx.doi.org/10.1111/j.1574-6968.1997.tb10355.x
Suzuki S., Katagiri S., Hideaki, 1996, Mutants with altered sensitivity to a CaM antagonist affect the circadian clock in Neurospora crassa, Genectics, 143:1175-1180
Suzuki C., 2001, Immunochemical and mutational analysis of P-type ATPase Spf1p involved in the yeast secretory pathway, Biosci Biotecnol Biochem, 65:2405-2411
http://dx.doi.org/10.1271/bbb.65.2405
Tamuli R., Kumar R., Deka R., 2011, Cellular roles of neuronal calcium sensor-1 and calcium/calmodulin-dependent kinases in fungi, J Basic Microbiol, 51:120-128
http://dx.doi.org/10.1002/jobm.201000184
Tamuli R., Kumar R., Srivastava D.A., Deka R., 2013, Calcium signaling. In: Kasbekar DP, McCluskey K (eds) Neurospora: genomics and molecular biology, Caister Academic Press, Norfolk, pp 35-57
Tamura K., Peterson D., Peterson N., Stecher G., et al., 2011, MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods, Mol Biol Evol, 28:2731-2739
http://dx.doi.org/10.1093/molbev/msr121
Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., et al., 1997, The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools, Nucleic Acids Res, 25:4876-4882
http://dx.doi.org/10.1093/nar/25.24.4876
Yang Q., Borkovich K.A., 1999, Mutational activation of a Gαi causes uncontrolled proliferation of aerial hyphae and increased sensitivity to heat and oxidative stress in Neurospora crassa, Genetics, 151:107-117
Zalokar M., 1954, Studies on biosynthesis of carotenoids in Neurospora crassa, Arch Biochem Biophys, 50:71-80
http://dx.doi.org/10.1016/0003-9861(54)90010-7
. PDF(1743KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Vijya Laxmi
. Ranjan Tamuli
Related articles
. Calcium signaling
. Ca 2+ /cation ATPases
. Calmodulin
. Neurospora crassa
. nca-2
. thermotolerance
Tools
. Email to a friend
. Post a comment


