Construction of the Near-Isogenic Lines of the MI CMS Restorer Gene in Rapeseed (Brassica napus) and Allelic Analysis 
 Author
Author  Correspondence author
Correspondence author
Genomics and Applied Biology, 2011, Vol. 2, No. 6 doi: 10.5376/gab.2011.02.0006
Received: 20 Nov., 2011 Accepted: 09 Dec., 2011 Published: 16 Jan., 2012
Long et al., 2011, Construction of the Near-Isogenic Lines of the MI CMS Restorer Gene in Rapeseed (Brassica napus) and Allelic Analysis, Genomics and Applied Biology, 2011, Vol.2, No.6 34-41 (doi: 10.3969/gab.2011.02.0006)
In this research, we employed Ning R7, an elite restorer for MI CMS, as the restorer gene donor to develop restorer gene candidate near-isogenic lines (NILs) by using three elite germplasms, T78, T84 and ZS9, as recurrent parents based on the advanced backcross approach. The characteristics of agronomic traits were phenotyped at the maturity stage and genetic similarity was calculated based on the SSR data. The results showed most agronomic traits of the candidates had quite similar with that of recurrent parents besides the trait of plant height and the height of the initiate branch based on t-test. Further, in order to figure out the similarity of the genetic background, we used 75 SSR markers to generate total 157 SSR loci in the 21 candidate NILs. The average genetic similarity coefficient of the candidate NIL background reached 0.86, 0.92 and 0.98 corresponding to their own recurrent parent, respectively. With the comprehensive assessment, three candidate NILs, wh13, wh59 and wh118, were chosen as potential restorer NILs, which might be used in our future breeding program.
Cytoplasmic male sterility system (CMS) would be the most important way for heterosis utilization at the present in China. MI CMS system of Brassica napus type is a kind of three line breeding system developed by Mr. Shou-Zhong Fu of Jiangsu Academy of Agricultural Sciences with independent intellectual property rights (Fu et al., 1989). Series of new varieties of hybrid rapeseed, such as Ningza No1, Ningza No3 Suyou No5, Ningza No 15, Ningza No19 and Ningza No21 were developed and released by using this breeding system. The utilization of hybrid rapeseed in China has been placed on the front ranks in the world (Fu, 1999), but the genetic mechanism was just beginning. This system of MI CMS, the restoration of fertility was controlled by the single dominant gene (Zhang et al., 2000), thus restoring gene should be one of the most worthy to study in genetics. Scholars from countries were very keen on studying restoring genes of various Brassica napus type hybrid system by using BC1 and or F2 populations and DNA Bulk approach to obtain molecular markers linked to restoring genes of Pol CMS, Shaan 2A CMS and MI CMS systems (Jean et al., 1997, 1998; Zeng et al., 2009; Liu et al., 2007; Zhang et al., 2004; Zhang, 2005). It was obvious that the researchers focused on using early separating generations (such as BC1, F2) to map and clone the genes, however, there was very little using near-isogenic lines of the restoring gene to study the genes. Actually, the near-isogenic lines were less utilized in the research due to time consuming and low efficiency to building near-isogenic lines. However, it became an unique tool for studying the dominant genes of important traits in crops because of near-isogenic lines with a single genetic background, which might have more effective to analyze genetic effects, gene fine mapping and cloning, etc. Definitely, near-isogenic line would be the valuable materials in the fields of molecular biology and genetics, which were widely used in rice, corn and other crops to have obtained the molecular markers close linked to the target traits as well as to have cloned the target genes (Toshiyuki et al., 2003; Zhang et al., 2006).
In the building process of near-isogenic lines, generally, each generation used to be done by phenotypic selection. After multi-generation crossing, there is little interaction between traits and environments, some negative factors such as the linkages of the donor gene fragments with target genes can make the bias to select the offspring lines, which will affect the quality of building the near-isogenic lines. Thus, after the near-isogenic lines built, it is very necessary to assess the allelism of the candidates of near-isogenic lines. In the early, traditional methods commonly phenotypic trait or traits related to the studied objects were used to evaluate the near-isogenic lines, whereas the molecular markers rapidly developed in recent years have been used for detecting the allelism (Shen, 2004; Ma, et al., 2007; Liu, 2006; Liu, et al., 2008; Xu, et al., 2008).
The near-isogenic lines of the restore genes in MI CMS rapeseed hybrid system is a powerful tool for tagging and cloning the restore genes. Since 2002 the authors had started to build the near-isogenic lines of restore genes of MI CMS system, we obtained the near-isogenic lines with three different genetic backgrounds through crossing with three genetic background elite germplasms and selecting in the multi-generation backcross. Although there are lots of approaches for the near-isogenic lines, it is almost blank using multiple methods to simultaneously detect the allelism. In this study we carried out the genetic similarity of some candidates of near-isogenic lines of restore genes derived from three different genetic backgrounds by using rapeseed (AACC) SSR, and assessed the allelism of candidates to their donor’s background by combining agronomic trait analysis of candidate lines. We attempted to obtain the best near-isogenic lines of restore genes to lay a solid foundation for tagging and cloning the restore genes in MI CMS system.
1 Results and Analysis
1.1 Building the near-isogenic lines of the fertile restoring genes in MI CMS
In this research we employed Ning R7 as the source of restoring gene, T78, T84 and ZS9 as the donors of genetic backgrounds. Since 2002, we adopted the continuous backcrossing procedures method to substitute the genetic background to select far more similar individuals with donor’s phenotypes for further backcross. Up to 2008, we reached BC5F1 generation with three different genetic backgrounds. In the beginning of the BC5F1 generation, we adopted the strategy by combining the backcrossing and selfing to continually increase the substitution rate of the genetic background. In the generations of BC6F1 and BC5F2 generations, fertility segregation was investigated in the flowering stage to eliminate the lines that had fertility segregating. In BC5F3 generations, lines with full fertile individuals were selected to be as tentative candidates of near-isogenic lines with homozygous restore genes. In 2010, seven, six and eight candidate NILs related to three genetic backgrounds respectively were come out (Table 1). The detailed building procedures were shown in Figure 1.
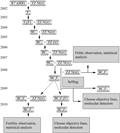 Figure 1 The building route of the NILs for restorer genes |
 Table 1 The values of agronomic traits and t values of the candidate NILs with three genetic backgrounds (t0.05ï¼2.26, t0.01ï¼3.25, n=10) |
1.2 Phenotyping agronomic traits of NIL candidates
The variance analysis of agronomic traits among the NIL candidates and three genetic background donors was carried out by using t-test (Table 1). The results showed that the lines being differences in agronomic traits but having a higher genetic similarity with the donor happened in three genetic backgrounds. In the context of T78, the traits with the most significant differences were plant height and a number of the primary (first) branches, there were four lines with significant or very significant differences, whereas the traits of the lines with little difference were the thickness of stem and a number of silique of primary branches, which were no significant differences with the donor’s background of T78. The candidate lines with high genetic similarity with the donor of T78 were the lines of wh15, wh14 and wh13, comparing to the donor of T78, the line of wh15 only had significantly difference in the trait of branching point height, the wh13 in the traits of plant height and branching point height as well as the wh14 in the traits of the branching point height and the length principal inflorescence. In the context of T84, the traits with the most significant differences were plant height and branching point height, there were six lines with significant differences comparing to the parents, while there was no significant difference in the traits except the number of the second branch and the length of silique. The candidate lines with high genetic similarity with the donor of T84 were the lines of wh59 and wh78, which had significant difference in the trait of plant height and the branching point height comparing to the donor of T84. In the context of ZS9, the traits with the most significant differences were plant height and the branching point height, there were six and seven lines with significant differences, respectively, whereas the traits of the lines with little difference were the number of primary branch, the number of silique in principal inflorescence, the number of silique in primary branch, the length of silique, the number of seed per silique and 1000-seed weight. The candidate lines with high genetic similarity with the donor of ZS9 were the lines of wh118 and wh127 sequentially, which had significant difference in the trait of plant height and the branching point height comparing to the donor of ZS9. In terms of the comparison among three genetic background germplasms and their derived lines, there were eight traits with no significant differences in the candidates with T84 background, only two traits with no significant differences in the candidates with T78 background, while six traits with no significant differences in the candidates with ZS9 background. Therefore, the candidates of near-isogenic lines with T84 and ZS9 background had overall higher allelism than that of the candidates with T78 background based on the genetic similarity of agronomic traits.
1.3 Molecular identification of NIL candidates
NIL candidates derived from three genetic background of MI CMS system restore gene were identified by using 75 of SSR primer pairs. The results showed that total of 157 target bands in size from 100 bp to 500 bp were amplified by using 75 pairs of primers, each pair of primers amplified an average of 2.3 bands. There were 11 pairs, 4 pairs and 2 pairs respectively to generate only one polymorphic band in their genetic backgrounds. Figure 1 showed the banding pattern amplified in the three genetic backgrounds by the primer of H33. The results further indicated: (1) the same primer might come out with different amplification results in the same genetic background. Primers H33 generated clear 4 bands with different sizes in the T78 background, while only 2 bands in the T84 background as well as only one similar size band in ZS9 background. (2) Generally, all lines derived from the same background go to the same as the amplified results of each line, but the difference might happen, while the difference used as criteria for measuring the allelism of NILs. In the T78 background, all lines were consistent with the amplification results by H33, whereas in the T84 background, all lines had the same result of the band in size of about 200 bp, but the difference of the band in size about 450 bp. The banding patterns were quite different between lines of 9, 10, 15 and lines of 11, 12 and 13. In the ZS9 background, the banding pattern with a continuous bands in size amplified, which were the same in the lines of 16, 18, 22 and 17 while different in the line of 19, 20, 21, 23 and 24 (Figure 2).
| Figure 2 The banding pattern amplified by the primer H33 in 24 lines of three genetic backgrounds |
1.4 Clustering analysis of the NILs derived from the three genetic backgrounds
Based on the amplified results by molecular markers, the tree diagram was clustered by using cluster software (Figure 3). The results showed there were quite differences between the first and third genetic backgrounds. All lines with the same genetic background were grouped with the donor together. The correlation efficiency between the group T78 and ZS9 was 0.73, which indicated that two genetic backgrounds had high correlation. Both groups of T78 and ZS9 had 0.54 correlation efficiency with T84 group which presented quite difference. Thus we obtained two NIL lines with different genetic backgrounds fpr restore gene of MI CMS system in this research. Each candidate NILs in different background had correlation with parents, and the difference between the lines and donor was amplified a little. Genetic similarity efficiency of candidate lines from T78 group ranged from 84.50 to 91.47% with an average of 86.78%, the lines closest to T78 were wh3, wh13 and wh15. Candidate lines from T84 group had relatively high genetic similarity efficiency, ranged from 91.25 to 92.87% with an average of 92.38%, the lines closest to T84 were wh59, wh71 and wh77. Candidate lines from ZS9 group had the highest genetic similarity efficiency, ranged from 95.66% to 98.78% with an average of 98.14%, the lines closest to ZS9 were wh118, wh123 and wh127.
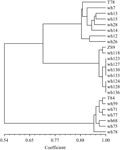 Figure 3 Cluster diagram of 24 NILs of restore genes based on the data of molecular markers |
2 Discussions
2.1 Analysis on effectiveness of backcross method to build the NILs of restorer gene rapeseed
Continuous backcross method is a classic approach to build near-isogenic lines of a crop trait, but for different traits, the difficulty of building strategy would be different, the building effect would be also quite different. For plant height and leaf shape, etc. these traits are visible to be easily selected in the backcross progeny with the naked eye (Zeng et al., 2006; Pu, 2001), so it is relatively easy to be built. Whereas for the traits of disease resistance and other traits that is invisible (Jin et al., 2007), the target traits are required to be examined in the backcross progeny before making a selection, and then advance the next backcross step, so it is more difficult to build NILs for such traits, which might take more time and be influenced by more factors. It might be more likely to choose the wrong target individual in the backcross resulting in failure building.
In this study, based on the principle of " co-existence of the sterile genes outside nucleus of cell and restorer genes inside nucleus of cell can exhibit the fertility" (Fu, 2000), we used Ning R7 as the female parent and sterile gene outside nucleus of cell as internal marker to simply select fertile individual to be as plant for continuing backcross, so that the restore gene will be guaranteed without loss, thus the NILs for the restore gene will be effectively built. In this study, the detections of fertility separation conducted in BC5F2 and BC6F1 generations have fully proved the existence of restorer genes. Overall, using this method to build NILs of restore gene has the following four advantages: (1) Significantly reducing the workload for detecting restorer gene in the offspring. Conventional approach need to conduct test cross for backcross progeny in order to prove the target line had the existence of restorer genes for advancing backcross, which is time-consuming and labor-intensive; (2) Completely guaranteeing the existence of restorer genes only by using the naked eye to determine the fertile phenotype. It does not only smartly apply the theory of classic three-line hybrid system, but also does not affect the substitution of the nucleus background; (3) On the premise of guaranteeing the existence of restore gene, it has a larger choice to choose offspring plants that are consistent with other phenotypic characteristics and the genetic background of donor, which would be more effective to substitute the genetic background; (4) The method developed in this study might be a relatively unique method for restorer breeding in three-lines hybrid breeding of rapeseed, which should be not only to reduce the workload for breeding restorer lines, but the targeted candidate of restore lines can reach 100% of recovery rate.
2.2 Comparison of detection methods for NIL’s allelism
The near-isogenic lines obtained by using successive backcross method might be failure because in the course of backcrossing in different years individual plant selection was influenced by the size of offspring population, cultivating methods, trait properties and other factors. Therefore, the allelism of candidate must be assessed. Traditional detection methods for allelism are generally to comparative agronomic traits, which would be working well for these visible traits. With the development of molecular biology, molecular markers have been adopted by more and more growing scholars. In this study, besides the use of sterile cytoplasm to tag the existence of the restoration genes, detecting the genetic background by using molecular markers and combined with agronomic phenotyping can identify much more excellent NILs, that is the procedure for NIL’s background detection by using both phenotype and genotype.
In this research we found that there is a certain difference in the lines selected by approach of phenotypic and genotypic selection. Phenotype is a kind of the external appearance of internal genotype of a line, which easily influenced by external environmental conditions during its growth and development. Whereas molecular detections mainly focus on identification of genetic alleles and loci, statistically speaking, which is a kind of sampling detection method that missing detection occurs. It is impossible to have all loci detected, so both of detection methods have some shortages. This study attempts to use two methods to evaluate the lines and mutually validate the data. Although the best targeted lines selected by two detection methods had some differences, the targeted lines were those lines identified by both approaches. Actually the results were similar. Finally, using two detection methods can validate the results from single detection to produce the better data. Definitely, selecting excellent Near-isogenic lines would be very important to fine mapping and gene cloning.
SSR, also known as microsatellite markers, presents a kind of variability based on the length of repeat nucleotide in the different locations of the living genome (Von Diethard, 1989). Since the SSR came out, with its stable reproducible and clear banding etc., SSR would be the dominant marker to tag the crop traits in rice, cotton and other major crops (Zhang, 2009; Wang, 2005). In recent years, there have been more distinctive types of molecular markers such as AFLP, SRAP and others (Vos et al., 1995; Li et al., 2001), Both of AFLP and SRAP have a good fit to detect the differences between the parents, but detecting the allelism of the homozygous lines is not better than that of SSR. AFLP procedures are much more complex including enzyme digestion than the procedures of SSR. The authors also tried to use the SRAP to detect allelism, which usually results in affecting the validity of scoring the data because of its random primer matching sites, more PCR products or multi-sites, or differences in the amount of amplification or less stability. Therefore, using SSR to detect the NIL’s allelism would occupy some good advantages.
2.3 Genetic similarity of restorer gene near-isogenic lines
Rapeseed (Brassica napus), a typical cross-pollinated crops, is allotetraploid with high homology between sub-genomic A and C. So in the breeding practice, it is very common that the stable line with several generation bagged-selfing still possible separate the different line with different phenotype of the original inbred lines. According to backcross protocol, the classical the background substitution rate should be 96.875% after five generations of backcrossing. In this study, the seven near-isogenic lines of T78 genetic background had only 86.78% of average genetic similarity, it is obvious far behind the theoretical value, which would be possible that the T78 had being varied in the selfing process. It is from the side to explain that building near-isogenic lines would be difficulty. Whereas near-isogenic lines of genetic similarity coefficient in the T84 and ZS9 background were up to 90% or more, especially the most successful to build near-isogenic lines of ZS9 background. In this study, using three different genetic background to simultaneous build NILs of restore gene can effectively avoid the individual selection error in the process of rapeseed background substitution, which might guarantee to obtain better near-isogenic lines, although the workload has increased much more, In addition, analysis of the genetic similarity coefficient showed that NILs from the background of T78 and ZS9 had high coefficient of genetic similarity, both of them had low genetic similarity coefficient with that of T84. Near-isogenic lines of restore gens built simultaneous in the large differences of genetic background become a good research resource for studying the functions of restore gene.
2.4 The prospects on utilization of rapeseed NILs of restore gene
The process for building NILs of restorer gene is relatively time-consuming and labor-intensive. Once the successful building of NILs is done, it has very important application values for studying restore gene and interactions between restore gene and sterile genes. First of all, NILs can be used to fine map the restore loci and clone the restore gene. Because of its almost the same genetic background, compared to other primary mapping populations such as F2 and BC1 etc., the mapping process may be relatively less of the interference. Secondly, using NILs of restorer gene can further crack the three-line hybrid system. All along, it is yet to be studied in-depth for influences of sterile gene of outside of cell nucleus on phenotypes of hybrids including yield traits, the restoring abilities to CMS restorers with different backgrounds, mechanism of influence of genes in outside cell nucleus (such as mitochondria genes, etc.) on the fertility of three-line hybrid system and so on. It would be very important meaning to these issues mentioned above to fully understand and apply the three-line hybrid system. The establishment of three NILs in this research will provide a good material supporting foundation for further studies of these issues mentioned above. Thirdly, the validation of restore gene functions. Restoring gene’s NILs are two kinds of the lines with or without restore gene in the same genetic backgrounds. It is the perfect material to validate the functions of restore gene by using the line without the restoring gene as targeted line being transferred the restore gene. Therefore, the restorer gene’s NILs should be the basis materials for studies, which has good prospects for future applications.
3 Materials and Methods
3.1 Rapeseed materials and SSR primers
Ning R7, the restorer line of Rapeseed MI CMS hybrid Ningza No 3, which the cytoplasm is sterile cytoplasm, and nucleus has homozygous restoring gene, used as the source of restorer gene.
T78, T84 and ZS9, used as genetic background donors, are Brassica napus germplasms with large differences from different genetic backgrounds and comprehensive excellent agronomic traits of Brassica, which could not restore the fertility of CMS by identification of restor-maitaining relationship. These materials mentioned above were provided by Research group of rapeseed at Institute of industrial crops of Jiangsu Academy of Agricultural Sciences.
Total of 75 SSR primer pairs were chosen from these have the high polymorphism among Brassica napus varieties, parts of primers kindly provide by Research group of rapeseed of Huazhong Agricultural University, others were downloaded from the website of www.brassica.info. All of primers were synthesized by Shanghai Yinjun Biotechnology Company.
3.2 Phenotyping and analysis of agronomic traits
Ten individuals with similar plant type were chosen to measure the agronomic and economic characters prior to eliminate both ends of individual of the row before harvest at the stage of rapeseed maturity. Taking the average as measured values. Comparative analysis of the trait values between the genetic background and the candidate lines were carried out by using t test, when the character value went to significant difference the corresponding t-values will be marked in the upper right corner.
3.3 Extraction of genomic DNA of rapeseed
We adopted the CTAB methods developed by Li et al (1994) with slightly modification to extract genomic DNA from rapeseed leaves. The bulk of leaves mixed from five similar plant type individuals of each line were ground. The concentration of dissolved DNA was examined by 1% agarose gel and adjusted the concentration of each line.
3.4 Screening of SSR primers and amplification
Using 75 pairs of SSR primers, genomic DNA of NILs of T78, T84 and ZS9 were amplified to screen the primer pairs with good polymorphism, appropriate band size and stable amplification. PCR procedure was performed following the reference of Long et al (2009). PCR products were separated by 8% polyacrylamide gel electrophoresis with 20 V/cm until the front of the Loading buffer migrated to the other edge, and then silver staining was carried out.
3.5 Band scoring, statistics and cluster analysis
Bands amplified by SSR markers with clear, sharp and heavy band were scored and calculated statistically. Band was assigned to be 1, and no band was assigned to be 0 based on the relevant position. Unclear band was assigned to be “-”. Dendrogram was drawn by cluster analysis using NTSYS software.
Author’s contributions
WHL and MLH are the persons who carried out this experiment; JQG, HMP, SC, and JFZ participated in some in lab work and involved the data analysis and in field work; CKQ conceived the project and designed the experiments as well as wrote and revised manuscript. All authors had read and agreed the final text.
Acknowledgements
This research is jointly sponsored by the National 863 Project (No.2011AA10A104), Supporting program of Jiansu province (BE2008369) and Independent Agricultural Innovation Program of Jiangsu Province (cx(09)636) Authors thank for two anonymous reviewers with their critical comments. In this paper we mentioned some chemical and reagent suppliers and sequencing service providers, that doesn't mean we would like to recommend or endorse the production of theirs.
Reference
Fu S.Z., Qi C.K., and Tang J.H., 1989, Breeding of cytoplasmic male sterile line MI CMS in Brassica napus L., Zuowu Xuebao (Acta Agronomica Sinica), 15(4): 305-309
Fu T.D., 1999, Rapeseed production and variety improvement in China, Huazhong Nongye Daxue Xuebao (Journal of Huazhong Agriculture University), 18(6): 501-504
Fu T.D., 2000, Breeding and utilization of rapeseed hybrid, Hubei Science and Technology Press, Wuhan, China, pp.11-20
Jean M., Brown G.G., and Landry B.S., 1997, Genetic mapping of nuclear fertility restorer genes for the ‘Polima’ cytoplasmic male sterility in canola (Brassica napus L.) using DNA markers. Theoretical and Applied Genetics, 95(3): 321-328
http://dx.doi.org/10.1007/s001220050566
Jean M., Brown G.G., and Landry B.S., 1998, Targeted mapping approaches to identify DNA markers linked to the Rfp1 restorer gene for the ‘Polima’ CMS of canola (Brassica napus L.). Theoretical and Applied Genetics, 97(3): 431-438
http://dx.doi.org/10.1007/s001220050913
Jin X.W., Wang C.L., Yang Q., Jiang Q.X., Fan Y.L., Liu G.C., and Zhao K.J., 2007, Breeding of Near-Isogenic line CBB30 and molecular mapping of xa30(t), a new resistance gene to bacterial blight in rice, Zhongguo Nongye Kexue (Scientia Agricultura Sinica), 40(6): 1094-1100
Li G., and Quiros C.F., 2001, Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica, Theoretical and Applied Genetics, 103(2): 455-461
http://dx.doi.org/10.1007/s001220100570
Li J., and Shen B.Z, 1994, An effective procedure for extractive total DNA in rape, Huazhong Nongye Daxue Xuebao (Journal of Huazhong Agriculture University), 13(5): 521-523
Liu L.K., Hou N., Liu J.C., Liu G.Q., and Liu C.G., 2006, Screening of the near-isogenic lines (NILs) of fertility restorer genes for D^2-type cytoplasmic male sterility (CMS) in wheat and molecular characterization of the genetic backgrounds, Mailei Zuowu Xuebao (Journal of Triticeae crops), 26(5): 1-4
Liu P.W., Li Y., He Q.B., and Yang G.S., 2007, Identification of genetic markers for the pol CMS restorer gene in Brassica napus L., Zhongguo Youliao Zuowu Xuebao (Chinese journal of oil crop science), 29(1): 14-19
Liu Y.H., Yan H.F., Yang W.X., Li X., Li Y.N., Meng Q.F., Zhang L.R., Liu D.Q., and Zhang T., 2008, Molecular diversity of 23 wheat leaf rust resistance near-isogenic lines determined by sequence-related amplified polymorphism, Zhongguo Nongye Kexue (Scientia Agricultura Sinica), 41(5), 1333-1340
Long W.H., Gao J.Q., Pu H.M., Qi C.K., Zhang J.F., and Chen S, 2009, Molecular fingerprints construction of the new rapeseed variety ningza No.15, Jiangsu Nongye Xuebao (Jiangsu Journal of Agriculture Science), 25(6): 1238-1242
Ma S.C., Zhang G.S., and Niu N., 2007, Breeding of near-isogenic lines of multi-ovary character in wheat and their genetic background evaluation, He Nongxue Bao (Acta Agriculture Nucleatae Sinica), 21(6): 585-588
Pu H.M., Fu S.Z., Qi C.K., Zhang J.F., Wu Y.M., Gao J.Q., and Chen X.J., 2001, Inheritance of divided leaf trait of rapeseed (Brassica napus) and application in hybrid breeding, Zhongguo Youliao Zuowu Xuebao (Chinese journal of oil crop science), 23(3): 60-61
Shen S.Q., Zeng Y.W., and Pu X.Y., 2004, Evaluation of near-isogenic lines of cold tolerance at booting stage of rice, Xinan Nongye Xuebao (Southwest China Journal of Agriculture Sciences), 17(S): 18-23
Toshiyuki K., Toshio Y., Naoki T., Masakazu K., and Hideko M., 2003, Fine genetic mapping of the nuclear gene, Rf-1, that restores the bt-type cytoplasmic male sterility in rice (Oryza sativa L.) by PCR-based markers, Euphytica, 129(2): 241-247
http://dx.doi.org/10.1023/A:1021915611210
Von Diethard T., 1989, Hypervariabflity of simple sequences as a general source for polymorphic DNA markers, Nucleic Acids Research, 17(16): 6463-6471
http://dx.doi.org/10.1093/nar/17.16.6463 PMid:2780284 PMCid:318341
Vos P., Hogers R., Bleeker M., Reijans M., van de Lee T., Hornes M., Friters A., Pot J., Paleman J., Kuiper M., and Zabeau M., 1995, AFLP: a new technique for DNA fingerprinting, Nucleic Acids Research, 23(21): 4407-4414
http://dx.doi.org/10.1093/nar/23.21.4407 PMid:7501463 PMCid:307397
Wang S.H., and Du X.M., 2005, SSR fingerprinting analysis on distinct mutants of fiber development in gossypium hisutum, Zhongguo Nongye Kexue (Scientia Agricultura Sinica), 38(10): 2139-2146
Xu M.H., Xu S.Z., Li X.X., Lv J.G., Peng D.X., and Wang Z.M., 2008, Development of monoecious NILs and assessment of the Near-Isogenic level in melon, Fenzi Zhiwu Yuzhong (Moleculer Plant Breeding), 6(2): 302-308
Zeng F.Q., Yi B., Tu J.X., and Fu T.D., 2009, Identification of AFLP and SCAR markers linked to the male fertility restorer gene of pol CMS (Brassica napus L.), Euphytica, 165(2): 363-369
http://dx.doi.org/10.1007/s10681-008-9799-x
Zeng X., Zhu L., Chen Y., Qi L., Pu Y., Wen J., Yi B., Shen J., Ma C., Tu J., and Fu T., 2010, Identification, fine mapping and characterization of a dwarf mutant (bnaC. dwf) in Brassica napus, Theoretical and Applied Genetics, 122(2): 421-428
http://dx.doi.org/10.1007/s00122-010-1457-8 PMid:20878141
Zhang J.F., Fu S.Z., and Gai J.Y., 2000, Inheritance of fertility of cytoplasmic male sterile system MI CMS in rapeseed (Brassica napus L.), Zhongguo Youliao Zuowu Xuebao (Chinese journal of oil crop science), 22(1): 14-18
Zhang J.F., Fu S.Z., Qi C.K., Pu H.M., Chen X.J., and Gao J.Q., 2005, Identification of RAPD markers linked to the fertility restore gene of MI CMS in rapeseed (Brassica napus), Zhongguo Youliao Zuowu Xuebao (Chinese journal of oil crop science), 27(2): 1-4
Zhang M.L., Lin B.G., Zhang X.Y., Cui H.R., and Xia Y.W., 2004, Study on inheritance and restoring gene linked RAPD markers of cytoplasmic male sterility of Shan 2A (Brassica napus L.), He nongxue bao (Acta Agriculture Nucleatae Sinica), 18(6): 427-430
Zhang Y., Li Y.F., Xie R., Yang Z.L., Zhong B.Q., Shen F.C., Tan Z.J., and He G.H., 2009, Development of NILs with Cl-gene of rice Restorer and Evaluation on the Near-isogenic level Clustered spikelets, Zuowu Xuebao (Acta Agronomica Sinica), 25(6): 397-401
Zhang Z.F., Wang Y., and Zheng Y.L., 2006, AFLP and PCR-based markers linked to Rf3, a fertility restorer gene for S cytoplasmic male sterility in maize, Molecular Genetics and Genomics, 276(2): 162-169
http://dx.doi.org/10.1007/s00438-006-0131-y PMid:16705419
. PDF(403KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Weihua Long
. Maolong Hu
. Jianqin Gao
. Huiming Pu
. Song Chen
. Jiefu Zhang
. Cunkou Qi
Related articles
. Rapeseed ( Brassica napus L.)
. CMS
. Restorer gene
. Near-Isogenic Lines (NIL)
. Allelic analysis
Tools
. Email to a friend
. Post a comment


