2. Department of Applied Biological Sciences, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo, 113-8657, Japan
 Author
Author  Correspondence author
Correspondence author
Genomics and Applied Biology, 2011, Vol. 2, No. 3 doi: 10.5376/gab.2011.02.0003
Received: 06 Sep., 2011 Accepted: 20 Oct., 2011 Published: 03 Nov., 2011
Bu et al., 2011, Research Progress of Ammonium Transporter in Rice Plants, Genomics and Applied Biology, 2011, Vol. 2, No. 3 (DOI: 10.5376/gab.2011.02.0003)
Plant taken up from the soil have two inorganic forms, ammonium ion (NH4+) and nitrate ion (NO3-), but rice grows in flooded paddy fields and takes up ammonium as the preferred nitrogen (N) source. Ammonium uptake is predominantly mediated by ammonium transporters in the plasm membrane. Ammonium transporter genes (AMTs) have been cloned and identified in many organisms.So far, total 12 AMT proteins identified in rice, containing 10~11 transmembrane domains, except OsAMT4 and OsAMT5.2. In this review, we summarized the gene expression and regulation of ammonium transporters under different nitrogen conditions. Effective NH4+ uptake can provide a favorable protection for improving of agricultural production.
Nitrogen is one of the essential macronutrient for rice growth and one of the main factors to be considered for developing, a high-yielding rice cultivar and taken up ammonium as their preferred N source, and also ammonium tolerance. Rice grown in paddy field predominantly utilizes ammonium during most of the growing period. The first step in nitrogen assimilation is the uptake nitrate and ammonium into root cells from the soil solution. Root uptake of nitrate can be stored in the root cell vacuoles or through the xylem to the shoot, also can be reduced to nitrite by nitrate reductase, then enter the plastid reduced to ammonia ion by nitrite reductase, rapid assimilated into amide residue of glutamine by the couple reaction of glutamine synthetase and glutamate synthase. But ammonium is important from the external environment, including both the rhizosphere and atmosphere via ammonium transporters in the plasm membrane of the root cells and leaf cells. Ammonium entering the cell is assimilated either in the cytoplasm via glutamine synthetaseor in the plastids and possibly mitochondria following transport into these organelles, and ammonium may also enter the vacuole where it is stored temporarily. Ammonium can also be generated from N2 by root nodule cells. Most of this ammonium is transferred to the plant cytoplasm where it is assimilated into glutamine by GS/GOGAT cycle for plant growth. Since ammonium assimilation requires less energy than that of nitrate, ammonium is the preferential form of nitrogen uptake when plants are subjected to nitrogen deficiency. Using positron- emitting 13N-labeled ammonium (13NH4+) to monitor the movement from root to shoot in rice plant, the plant was cultured under normal conditions, 13NH4+ supplied to roots was taken up, and nitrogen deficiency enhanced 13N translocation, this suggested ammonium assimilation actively on ammonium transporters or glutamine synthetases (Kiyomiya et al., 2001).
Ammonium uptake is predominantly mediated by ammonium transport systems at the root plasma membrane: a high-affinity transport system (HATS) and a low-affinity transport system (LATS), and constitute a multi-gene family, have been isolated in several plant speciesFingure.1.Phylogenetic tree analysis of the ammonium transporter genes cloned in plant species (Figure 1) (Luo and Liu, 2009), including Arabidopsis thaliana (AtAMT1;1-AtAMT1;5, AtAMT2) (Kaiser et al., 2002; Sohlenkamp et al., 2002), Oryza sativa (OsAMT1;1-OsAMT1;4, OsAMT2; 1-OsAMT2;3, OsAMT3;1-OsAMT3;3, OsAMT4 and OsAMT5) (Suenaga et al., 2003; Sonoda et al., 2003a), Lycopersicon esculentum (LeAMT1;1-LeAMT1;3, LeAMT2) (Lauter et al., 1996; Von Wiren et al., 2000); Triticum aestivum (TaAMT1;1-TaAMT1;3, TaAMT2; 1) (Jahn et al., 2004); Lotus japonicas (LjAMT1;1, LjAMT2;1) (Salvemini et al., 2001; Simon-Rosin et al., 2003); Brassica napus (BnAMT1;2) (Pearson et al., 2002). These results show that the ammonium transporter genes are widely exist in many plants which not only uptake of ammonium ion under a wide concentration range, may also plays different effect of regulation in the specificity tissue or organ.
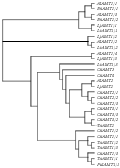 Figure 1 Phylogenetic tree analysis of the ammonium transporter genes cloned in plant species (Luo and Liu, 2009) |
1 The AMT family of ammonium transporters in plants
The first ammonium transporter AtAMT1.1 was isolated from the model plant Arabidopsis thaliana by growth complementation of a yeast mutant defective ammonium uptake (Ninnemann et al., 1994), encodes a high-affinity transporter and energy-dependent with a Km value of <0.5 µmol/L, belong to AMT subfamily and is expressed in roots and shoot (Gazzarrini et al., 1999). The low affinity system is non-saturable with a Km in the millimolar concentration range (D'Apuzzo et al., 2004), like AtAMT2 was sequence-wise more closely related to the ammonium transporters Mep1, 2 and 3 from Saccharomyces cerevisiae and with AmtB from E. coli, forming, together with other prokaryotic homologues, the MEP subfamily (Ludewig et al., 2001). So far, the third set of homologous sequences does not contain any plant protein, mainly includes human and animal Rheus blood group antigens, thus forming the Rh subfamily of ammonium transporters. With the genome sequencing of the graminaceous model plant rice, so far, twelve putative OsAMT genes were identified, listed in Table 1 (Li et al., 2009a), Of these genes, 10 were previously identified by Suenaga et al and subdivide into four clades, OsAMT1 to OsAMT4. The amino acid sequences of the OsAMT1;1, OsAMT1;2 andOsAMT1;3 share high sequence similarity to each other and are very dissimilar to the other OsAMT families. The phylogenetic analysis showed that they could be divided into two subfamilies, AMT1 and AMT2. Except for the family OsAMT4 which contain only one member OsAMT4.1, each of the three families contain three members, OsAMT1 (OsAMT1.1, OsAMT1.2, OsAMT1.3), OsAMT2 (OsAMT2.1, OsAMT2.2, OsAMT2.3), OsAMT3 (OsAMT3.1, OsAMT3.2, OsAMT3.3). At present the OsAMT5.1 also identified (Deng et al., 2007). The 12 AMT genes distribute on rice chromosomes 1 to 5, 11 and 12. Chromosomes 1 and 2 contained three genes each, chromosome 3 contained two genes, and chromosomes 4, 5, 11 and 12 contained one gene each, and have 10~11 transmembrane regions with an extracellular N-terminus and an intracellular C- terminus.
.png) Table 1 List of predicted AMT genes in rice (Li et al., 2009b) |
2 The functional and expression of ammonium transporters
Using heterologous expression in yeast to determine whether the protein encodes a functional ammonium transporter or not. The yeast strain 31019b is deleted in three endogenous ammonium transporter genes (MEP1, MEP2, MEP3) and unable to grow on medium containing less than 5 mmol/L NH4+ as the sole nitrogen source. The results obviously shown that OsAMT1.1, OsAMT1.2, OsAMT2.1 and OsAMT5.1 all function domains as ammonium transporters (Suenaga et al., 2003; Deng et al., 2007). Studies on expression and regulations of ammonium transporter genes in rice have been focused on the three genes of OsAMT1 family, displayed different expression patterns in response to change in N levels. Northern blot analysis showed a distinct expression pattern for the three genes; OsAMT1.1 expression was constitutive in shoots and promoted by ammonium in roots; OsAMT1.2 expression was root-specific and ammonium-inducible; OsAMT1.3 was also expressed specifically in roots but was repressed by nitrogen, but expression levels of OsAMT1; 3 were quite low. Over-expression of plant membrane transporters have previously been used to determine whether they are the rate limiting factors in mineral uptake and compartmentalization. So over-expression of the OsAMT1.1 can increases ammonium uptake and content, but impairs growth and development of plants under high ammonium nutrition (Hoque et al., 2006). In situ mRNA detection revealed that OsAMT1;2 is expressed in the central cylinder and cell surface of root tips (Sonoda et al., 2003a). Thus, OsAMT1.2 may be involved in two functions, which are ammonium uptake from the soil solution and the uptake and retrieval of ammonium in the vascular system. Also, point mutant of OsAMT1.2 by using gene splicing by overlap extension method that anticipant mutant genes were obtained (Li et al., 2009b), which is significant to study structurally and functional characteristic of gene for ammonium transporters in rice.
In higher plant, few reported about the genes of AMT2 family, most members of the AMT2 family are low-affinity transporter. In fact, AMT2 is distantly related to plant AMT1, but more closely related to ammonium transporters from prokaryotes. It is unable to transport ammonium analogue, methylamine. In rice, AMT2 subfamily genes were up regulated by nitrogen deprivation. Nitrogen conditions regulate AMT2 gene transcripts differently. The OsAMT2;1 encoded a functional ammonium transporter which was being constitutively expressed in roots and shoots irrespective of the nitrogen supply (Suenaga et al., 2003). OsAMT2.2 genes were detectable both in root and shoot by real-time quantitative pcr (Li and Shi, 2006), and expressed highly in short time after transfer to various nitrogen from no nitrogen nutrition, and then were regulated feedback, but had no distinct difference between the root and shoot. Other members of function and expression have not been reported. So in future work, AMT2 family study should be comprehensive explanation, in spite of homology, molecular structure and expression characteristics among the two families have obvious differences, but they are play an important role in ammonium uptake, it will be effective utilization of nitrogen for plant.
3 Glutamine synthetase plays an important role involved in ammonium assimilation
Nitrogen assimilation plays an important role in the growth and development of plants. The first enzymes involved in ammonium assimilation are glutamine synthetase (GS). The expression of OsAMT1 are closely relation to the endogenesis glutamate, the ammonium effect were prevented by methionine sulfoximine, an inhibitor of glutamine synthetase. Moreover, glutamine rather than ammonium controls the expression of ammonium transporter genes in rice (Sonoda et al., 2003b). Most of the NH4+ taken up by the roots can be assimilated within the roots. There are two glutamine synthetases in rice, GS1 and GS2, GS1 is important for normal growth and development and GS2 for the photorespiratory metabolism of N in chloroplasts. Specifically OsGS1.1 is critical for normal growth and grain filling. Using mutant lacking OsGS1.1 show that this enzyme is control the global metabolic network when plant growing under the ammonium as the nitrogen source (Kusano et al., 2011) and NADH-GOGAT1 is important for primary ammonium assimilation in roots at the seedling stage (Tamura et al., 2010). GS-over-expression in rice plant may be functioning in vegetative growth; higher GS activities increased the plant metabolic level and may accelerate plant growth, which could accelerate the leaf senescence, break the nitrogen compounds translocation and re-assimilation in the plant developmental stages (Cai et al., 2009).
4 Future prospects
Nitrogen utilization within rice plants followed by NH4+ uptake and assimilation in the roots is a complex process that depends on many factors during the growth and development of plants. Of course, a number of activity genes and enzymes will be involved in the nitrogen utilization process, in many cases, the expression of each gene is regulated in a cell type, but current knowledge is limited. Reverse genetics is a powerful approach to obtain conclusive evidence on the function of the corresponding gene products, also enzymes involved in metabolic of nitrogen assimilation pathway can be characterized by reverse genetics. But now it is very difficult to identify target genes that are involved in regulation, unlike enzymes in metabolic pathway, so an approach of quantitative trait loci (QTLs) analysis could be one way to isolate regulatory genes. So in rice, the effect of QTLs was the further research to confirmed target genes controlling uptake, assimilation, and metabolism of nitrogen, as well as nitrogen use efficiency.
Authors' contributions
In this paper, Keisuke Nemoto and Shenkui Liu are responsible for overall planning and management. Collecting data and article writing were completed by Yuanyuan Bu and Tetsuo Takano.
Acknowledgements
This work was supported by the Heilongjiang Provincial Program for Distinguished Young Scholars (JC200609) and State Forestry Administration 948 Program of PR China (No. 2008429) to Shenkui Liu.
References
Cai H., Zhou Y., Xiao J., Li X., Zhang Q., and Lian X., 2009, Over-expressed glutamine synthetase gene modifies nitrogen metabolism and abiotic stress responses in rice, Plant Cell Rep., 28(3): 527-537 http://dx.doi.org/10.1007/s00299-008-0665-z PMid:19123004
D'Apuzzo E., Rogato A., Simon-Rosin U., El Alaoui H., Barbulova A., Betti M., Dimou M., Katinakis P., Marquez A., Marini A.M., Udvardi M.K., and Chiurazzi M., 2004, Characterization of three functional high-affinity ammonium transporters in Lotus japonicus with differential transcriptional regulation and spatial expression. Plant Physiol., 134(4): 1763-1774 http://dx.doi.org/10.1104/pp.103.034322 PMid:15075393 PMCid:419849
Deng R.L., Gu J.T., Lu W.J., Xu H.R., Cao Y.F., Xiao K., 2007, Characterization, function and expression analysis of ammonium transporter gene OsAMT1.4 and OsAMT5 in rice (Oryza sativa), Sci. Agric. Sin., 40(11): 2395-2402
Gazzarrini S., Lejay L., Gojon A., Ninnemann O., FrommerW.B., Von WireÂn N., 1999, Three functional transporters for constitutive, diurnally regulated, and starvation-induced up take of ammonium into Arabidopsis roots, The Plant Cell, 11(5): 937-947 http://dx.doi.org/10.2307/3870826 PMid:10330477 PMCid:144234 http://dx.doi.org/10.1105/tpc.11.5.937
Hoque M.S., Masle J., Udvardi M.K., Ryan P.R., and Upadhyaya N.M., 2006, Over-expression of the rice OsAMT1-1 gene increases ammonium uptake and content, but impairs growth and development of plants under high ammonium nutrition, Functional Plant Biology, 33(2): 153-163 http://dx.doi.org/10.1071/FP05165
Jahn T.P., Mler A.L., Zeuthen T., Holm L.M., Klaerke D.A., Mohsin B., Kühlbrandt W., and Schjoerring J.K., 2004, Aquaporin homologues in plants and mammals transport ammonia, FEBS Lett., 574(1-3): 31-36 http://dx.doi.org/10.1016/j.febslet.2004.08.004 PMid:15358535
Kaiser B.N., Rawat S.R., Siddiqi M.Y., Masle J., and Glass A.D., 2002, Functional analysis of an Arabidopsis T-DNA “knockout” of the high-affinity NH4+ transporter AtAMT1;1, Plant Physiol., 130(3): 1263-1275 http://dx.doi.org/10.1104/pp.102.010843 PMid:12427993 PMCid:166647
Kiyomiya S., Nakanishi H., Uchida H., Tsuji A., Nishiyama S., Futatsubashi M., Tsukada H., Ishioka N.S., Watanabe S., Ito T., Mizuniwa C., Osa A., Matsuhashi S., Hashimoto S., Sekine T., and Mori S., 2001, Real time visualization of 13N-translocation in rice under different environmental conditions usingpositron emitting tracer imaging system, Plant Physiology, 125(4): 1743–1754 http://dx.doi.org/10.1104/pp.125.4.1743 PMid:11299355 PMCid:88831
Kusano M., Tabuchi M., Fukushima A., Funayama K., Diaz C., Kobayashi M., Hayashi N., Tsuchiya Y.N., Takahashi H., Kamata A., Yamaya T., and Saito K., 2011, Metabolomics data reveal a crucial role of cytosolic glutamine synthetase 1;1 in coordinating metabolic balance in rice, The Plant Journal, 66(3): 456-466 http://dx.doi.org/10.1111/j.1365-313X.2011.04506.x PMid:21255162
Lauter F.R., Ninnemann O., Bucher M., Riesmeier J.W., and Frommer W.B., 1996, Preferential expression of an ammonium transporter and of two putative nitrate transporters in root hairs of tomato, Proc. Natl. Acad. Sci., USA, 93(15): 8139-8144 http://dx.doi.org/10.1073/pnas.93.15.8139
Li B.Z., Li S.M., Shi W.M., and Su Y.H., 2009b, Point mutant of OsAMT112 by using gene splicing by overlap extension, Biotechnology Bulletin, 9: 69-72
Li B.Z., Merrick M., Li S.M., Li H.Y., Zhu S.W., Shi W.M., and Su Y.H., 2009a, Molecular basis and regulation of ammonium transporter in rice, Rice Science, 16(4): 314-322 http://dx.doi.org/10.1016/S1672-6308(08)60096-7
Li S.M., and Shi W.M., 2006, Quantitative characterization of nitrogen regulation of OsAMT1; 1, OsAMT1; 2, and OsAMT2; 2 expression in rice seedlings, Russian Journal of Plant Physiology, 53(6): 837-843 http://dx.doi.org/10.1134/S102144370606015X
Ludewig U., Von Wirén N., Rentsch D., and Frommer W.B., 2001, Rhesus factors and ammonium: a function in efflux? Genome Biology, 2: 1-5 http://dx.doi.org/10.1186/gb-2001-2-3-reviews1010
Luo Y.Y., and Liu S.K., 2009, Research progress of ammonium transporter in plants, Genomics and Applied Biology, 28(2): 373-379
Ninnemann O., Jauniaux J.C., and Frommer W.B., 1994, Identification of a high affinity NH4+ transporter from plants, EMBO Journal, 13(15): 3464-3471 PMid:8062823 PMCid:395249
Pearson J.N., Finnemann J. and Schjoerring J.K., 2002, Regulation of thehigh-affinity ammonium transporter (BnAMT1; 2) in the leaves of Brassica napus by nitrogen status, Plant Mol. Biol., 49(5): 483-490 http://dx.doi.org/10.1023/A:1015549115471 PMid:12090624
Salvemini F., Marini A.M., Riccio A., Patriarca E.J. and Chiurazzi M., 2001, Functional characterization of an ammonium transporter gene from Lotus japonicus, Gene, 270(1-2): 237-243 http://dx.doi.org/10.1016/S0378-1119(01)00470-X
Simon-Rosin U., Wood C., and Udvardi M.K., 2003, Molecular and cellular characterisation of LjAMT2;1, an ammonium transporter from the model legume Lotus japonicus, Plant Mol. Biol., 51(1): 99-108 http://dx.doi.org/10.1023/A:1020710222298 PMid:12602894
Sohlenkamp C., Wood C.C., Roeb G.W. and Udvardi M.K., 2002, Characterizationof Arabidopsis AtAMT2, a high-affinity ammonium transporter of the plasma membrane, Plant Physiol., 130(4): 1788-1796 http://dx.doi.org/10.1104/pp.008599 PMid:12481062 PMCid:166690
Sonoda Y., Ikeda A., Saiki S., Von Wirén N., Yamaya T. and Yamaguchi J., 2003a, Distinct expression and function of three ammonium transporter genes (OsAMT1;1–1;3) in rice, Plant Cell Physiol., 44(7): 726-734 http://dx.doi.org/10.1093/pcp/pcg083 PMid:12881500
Sonoda Y., Ikeda A., Saiki S., YamayaT., and Yamaguchi. J., 2003b, Feedback Regulation of the ammonium transporter gene family AMT1 by glutamine in rice, Plant Cell Physiol., 44(12): 1396-1402 http://dx.doi.org/10.1093/pcp/pcg169 PMid:14701935
Suenaga A., Moriya K., Sonoda Y., Ikeda A., Von Wirén N., Hayakawa T., Yamaguchi J., and Yamaya T., 2003, Constitutive expression of a novel-type ammonium transporter OsAMT2 in rice plants, Plant Cell Physiol., 44(2): 206-211 http://dx.doi.org/10.1093/pcp/pcg017 PMid:12610225
Tamura W., Hidaka Y., Tabuchi M., Kojima S., Hayakawa T., Sato T., Obara M., Kojima M., Sakakibara H., and Yamaya T., 2010, Reverse genetics approach to characterize a function of NADH-glutamate synthase 1 in rice plants, Amino Acids, 39(4): 1003-1012 http://dx.doi.org/10.1007/s00726-010-0531-5 PMid:20213442
Von Wirén N., Lauter F.R., Ninnemann O., Gillissen B., Walch-Liu P., Engels C., Jost W., and Frommer W.B., 2000, Differential regulation of three functional ammonium transporter genes by nitrogen in root hairs and by light in leaves of tomato, Plant J., 21(2): 167-175 http://dx.doi.org/10.1046/j.1365-313x.2000.00665.x PMid:10743657
. PDF(210KB)
. HTML
Associated material
. Readers' comments
Other articles by authors
. Yuanyuan Bu
. Tetsuo Takano
. Keisuke Nemoto
. Shenkui Liu
Related articles
. Rice
. Ammonium transporter
. Nitrogen assimilation
. Glutamine synthetase
Tools
. Email to a friend
. Post a comment


