2 Alkali Soil Natural Environmental Science Center (ASNESC), Northeast Forestry University, Harbin 150040, PR China
1 Asian Natural Environmental Science Center (ANESC), The University of Tokyo, Tokyo 188-0002, Japan
2 Alkali Soil Natural Environmental Science Center (ASNESC), Northeast Forestry University, Harbin 150040, PR China
1 Asian Natural Environmental Science Center (ANESC), The University of Tokyo, Tokyo 188-0002, Japan
2 Alkali Soil Natural Environmental Science Center (ASNESC), Northeast Forestry University, Harbin 150040, PR China
 Author
Author  Correspondence author
Correspondence author
Genomics and Applied Biology, 2013, Vol. 4, No. 1 doi: 10.5376/gab.2013.04.0001
Received: 18 Dec., 2012 Accepted: 24 Dec., 2012 Published: 30 Jan., 2013
Liu et al., 2013, Functional analysis of a type
AtPP2C52 is a plasma membrane type-2C protein phosphatase. In this study, AtPP2C52 promoter-GUS analysis revealed that AtPP2C52 gene was found in a broad expression spectrum with a higher level in the vascular and meristem. AtPP2C52 can interact with multiple proteins, including a proteasome maturation factor, UMP1, and a cysteine proteinase, RD21a, as well as the heterotrimeric G proteins β subunit, AGB1. By mutational analysis of AtPP2C52, it was identified that some residues were essential for AtPP2C52 to bind AGB1, UMP1 and RD21a, suggesting that these proteins should be potential substrates of AtPP2C52.
Protein phosphorylation regulates almost all aspect of cell life (Hunter, 1998; Cohen, 1997). In Arabidopsis genome, 112 protein phosphatases have been identified (Kerk et al., 2002). Protein phosphatases have been considered to be much more flexible enzymes, which have a larger number of substrates and present with overlapping activities (Lammers and Lavi, 2007).
Based on the substrate specificity and on the conservation of the catalytic domain, protein phosphatases were grouped into protein serine/ threonine (Ser/Thr) phosphatases and protein tyrosine phosphatases. Protein Ser/Thr phosphatases were classified into phosphoprotein phosphatases (PPPs) and metal-dependent protein phophatases (PPMs). The PPM family contains type-2C protein phosphatase (PP2C) subfamily and pyruvate dehydrogenase phosphatase (Cohen, 1997).
PP2Cs were found in all organisms, such as plants, bacteria, yeast, nematodes, insects, and mammals (Schweighofer et al., 2004). A distinguishing feature of PP2Cs is the requirement of bivalent cation (Mn2+ or Mg2+) for their catalytic activity. Meanwhile, the intracellular concentrations of Mg2+ and Mn2+ do not fluctuate substantially under physiological conditions. Therefore, the activities of PP2Cs may controlled predominantly by their tissue- or cell type-specific expression, subcellular compartmentalization, post-translational modification, or/and degradation (Lammers and Lavi, 2007).
In Arabidopsis, seventy-six PP2C genes were identified (Kerk et al., 2002). These genes were clustered into several groups, based on their sequence similarity (Schweighofer et al., 2004; Xue et al., 2008). Group A PP2C genes are annotated as negative regulators of the ABA response in plant (Hirayama and Shinozaki, 2007). On the other hand, SNF1-related protein kinase 2 (SnRK2) family, which is activated by ABA or osmotic stress, positively regulates the ABA response in various tissues (Mustilli et al., 2002; Yoshida et al., 2002; Fujii et al., 2007). Group A PP2Cs interacted physically with SnRK2s in various combinations, and efficiently inactivated SnRK2s via dephosphorylation of multiple Ser/Thr residues in the activation loop (Umezawa et al., 2009). In response to ABA, PP2C-dependent negative regulation can be canceled by ABA receptors, RCAR/PYRs, leading to activation of positive regulatory pathways (Ma et al., 2009; Park et al., 2009). Group A PP2Cs interacted physically with RCAR/PYRs. Members of Group B PP2C were shown to regulate stomata aperture, seed germination, abscisic acid inducible gene expression, and interact and inactivate mitogen-activated protein kinase (MAPK, Umbrasaite et al., 2010).
AtPP2C52 was clustered into Group E (Xue et al., 2008). The interaction between the heterotrimeric G proteins β subunit (AGB1) and AtPP2C52 has been confirmed by Y2H analysis and an in vitro pull-down assay in our previous work (Tsugama et al., 2012a). Here we proved that AtPP2C52 is expressed in almost all the plant organs with a higher level in the vascular and meristem. AtPP2C52 can interact with UMP1 and RD21a as well as AGB1.
1 Results
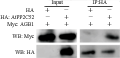
Figure 1 AtPP2C52 interacts with AGB1 in vitro
1.2 PAtPP2C52::GUS analysis

Figure 2 Temporal and spatial expression of AtPP2C52
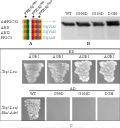
Figure 3 Site-directed mutations abolished the interaction between AtPP2C52 and AGB1
1.4 Potential substrates of AtPP2C52
The interaction of AtPP2C52 with either UMP1 or RD21a was confirmed by Y2H (Figure 4). AtPP2C52G99D and AtPP2C52DGH102-104ERN mutants failed to interact with UMP1 in Y2H (Figure 4A). G105D mutation did not affect the Y2H interaction between AtPP2C52 and UMP1 (Figure 4A). All of these mutations abolished the Y2H interaction between AtPP2C52 and RD21a (Figure 4B). The interactions were examined by a BiFC assay in Arabidopsis protoplast.

Figure 4 AtPP2C52 interacted with UMP1 and RD21a in Y2H
The ORFs of AGB1, UMP1 and RD21a were fused downstream of the nYFP and the ORF of AtPP2C52 was fused upstream of the cYFP. BiFC signals of nYFP-fused AGB1 and cYFP-fused AtPP2C52 were detected in the peripheral region of Arabidopsis mesophyll protoplasts (Figure 5) as previously described (Tsugama et al., 2012a). BiFC signals of nYFP-fused UMP1 and cYFP-fused AtPP2C52 were also detected in the peripheral region (Figure 5), suggesting that AtPP2C52 interacted with UMP1 in the plasma membrane. AtPP2C52 interacted with RD21a not only in the plasma membrane but also in the nucleus (Figure 5). These results suggest that RD21a and UMP1 are the potential substrates of AtPP2C52.
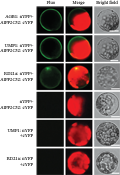 Figure 5 AtPP2C52 interacted with UMP1and RD21a in BiFC assays in Arabidopsis protoplasts |
2 Discussion
2.2 Mutational analysis of AtPP2C52
In conclusion, physical interactions between AtPP2C52 with UMP1, RA21a and AGB1 were confirmed. Mutational analysis shown that G99D, DGH102-104ERN and G105D are essential for AtPP2C52 to bind specific targets, such as UMP1, RA21a and AGB1.
3 Materials And Methods
3.2 Yeast two-hybrid (Y2H)
The point mutations of AtPP2C52 gene were generated by PCR using PrimeSTAR (TaKaRa) with wild type AtPP2C52 cDNA as template. For the point mutation of AtPP2C52G99D, PCR was performed using the following primer pair: G99D-Fw 5'-GTGACATTTTGTGATGTATTTGATGGTCATGGTCC-3' and AtPP2C52-Rv 5'-GAGTCGGATCCTCAAGTCTTCGATTTCTCTTC-3’ (BamHI site is underlined), generating 3'- terminus of AtPP2C52G99D. To generate 5’- terminus of AtPP2C52G99D, PCR was performed using the following primer pair: AtPP2C52-Fw 5'-GAGTCGAATTCATGGGGGGTTGTGTGTC-3' (EcoRI site is underlined) and G99D-Rv 5’-GACCATCAAATACACCACAAAATGTCACATCTTCAGAC-3’. Subsequently, the mixture of 3’- and 5’-terminus of AtPP2C52G99D was used as template for PCR using primer pair AtPP2C52-Fw and AtPP2C52-Rv, generating full-length AtPP2C52G99D. The PCR products of AtPP2C52G99D were digested by EcoRI and BamHI, and cloned into the EcoRI/BamHI site of pGADT7-rec, generating pGAD-AtPP2C52G99D. For the point mutation of AtPP2C52G99D, PCR was performed using the following primer pair: G105D-Fw 5’-GATGGTCATGATCCTTATGGCCATCTTGTTGCTCG-3’ and AtPP2C52-Rv, generating 3’-terminus of AtPP2C52G105D.
To generate 5'-terminus of AtPP2C52G105D, PCR was performed using the following primer pair: AtPP2C52-Fw and G105D-Rv 5’-GCCATAAGGATCATGACCATCAAATACACCACAAAATG-3'. Subsequently, the mixture of 3'- and 5'-terminus of AtPP2C52G105D was used as template for PCR using primer pair AtPP2C52-Fw and AtPP2C52-Rv, generating full-length AtPP2C52G105D. The PCR products of AtPP2C52G105D were digested by EcoRI and BamHI, and cloned into the EcoRI/BamHI site of pGADT7-rec, generating pGAD-AtPP2C52G105D.
For the point mutation of AtPP2C52DGH102-104ERN, PCR was performed using the following primer pair: DGH102-104ERN-Fw 5’-GGTGTATTTGAACGTAATGGTCCTTATGGCCATCTTG-3’ and AtPP2C52-Rv, generating 3’-terminus of AtPP2C52DGH102-104ERN. To generate 5’-terminus of AtPP2C52DGH102-104ERN, PCR was performed using the following primer pair: AtPP2C52-Fw and DGH102-104ERN-Rv 5’-CATAAGGACCATTACGTTCAAATACACCACAAAATGGC-3’. Subsequently, the mixture of 3’- and 5’-terminus of AtPP2C52 DGH102-104ERN was used as template for PCR using primer pair AtPP2C52-Fw and AtPP2C52-Rv, generating full-length AtPP2C52DGH102-104ERN. The PCR products of AtPP2C52 DGH102-104ERN were digested by EcoRI and BamHI, and cloned into the EcoRI/BamHI site of pGADT7-rec, generating pGAD-AtPP2C52DGH102-104ERN. These mutations were confirmed by sequencing and then used for yeast transformation.
Full-length cDNA of UMP1 (AT1G67250) and RD21a (AT1G47128) were ordered from ABRC and used as PCR templates for following plasmids construction. The ORF of UMP1 was amplified by PCR using the cDNA clone as template and the following primer pair: 5’-GAGTCGAATTCATGGAGTCTGAGAAAAAGATAGCTCATG-3’ (EcoRI site is underlined) and 5’-GAGTCGGATCCTTACATGAAACTTGGGTAAATCGG-3’ (BamHI site is underlined). The PCR products were digested by EcoRI and BamHI, and cloned into the EcoRI/BamHI site of pGADT7-rec, generating pGAD-UMP1. The ORF of RD21a was amplified by PCR using the cDNA clone as template and the following primer pair: 5’-GAGTCGAATTCATGGGGTTCCTTAAGCCAACCATGGC-3’ (EcoRI site is underlined) and 5’-GAGTCCCTAGGTTAGGCAATGTTCTTTCTGCCTTGTGACCAG-3’ (BamHI site is underlined). The PCR products were digested by EcoRI and BamHI, and cloned into the EcoRI/BamHI site of pGADT7-rec, generating pGAD-RD21a.
Yeast transformation using yeast strain AH109 and screening were performed by Matchmaker Gold Yeast Two-Hybrid System (Clontech). At least 5 colonies grown on the SD media lacking leucine and tryptophan (SD/-Leu/-Trp), were streaked on the SD/-Leu/-Trp and the SD media lacking leucine, tryptophan, adenine, and histidine (SD/-Trp/-Leu/-His/-Ade). Photos were taken after the yeasts were cultured for 3-5 days. The experiments were performed for three times.
3.3 In vitro Coimmunoprecipitation (Co-IP)
3.4 Bimolecular Fluorescence Complementation (BiFC) assay
3.5 Preparation of chimeric constructs
3.6 Plant transformation
3.7 GUS histochemical analysis
Author Contributions
Acknowledgements
http://dx.doi.org/10.1093/nar/12.22.8711
Clough S.J., and Bent A.F., 1998, Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana, Plant J., 16(6): 735-743
http://dx.doi.org/10.1046/j.1365-313x.1998.00343.x
http://dx.doi.org/10.1016/S0968-0004(97)01060-8
Das A.K., Helps N.R., Cohen P.T.W., and Barford D., 1996, Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 Ǻ resolution. EMBO J., 15(24): 6798-6809 PMCID: PMC452505
H., Verslues P.E., and Zhu J., 2007, Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis, Plant Cell, 19(2): 485-494
http://dx.doi.org/10.1105/tpc.106.048538
Hirayama T., and Shinozaki K., 2007, Perception and transduction of abscisic acid signals: Keys to the function of the versatile plant hormone ABA, Trends Plants Sci., 12(8): 343-35
http://dx.doi.org/10.1016/j.tplants.2007.06.013
Hunter T., 1998, The Croonian lecture 1997. The phosphorylation of proteins on tyrosine: its role in cell growth and disease, Philos. Trans. R. Soc. Lond. B Biol. Sci., 353(1368): 583-605 PMCID: PMC1692245
Kerk D., Bulgrien J., Smith D.W., Barsam B., Veretnik S., and Gribskov M., 2002, The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis, Plant Physiol., 129(2): 908-925
http://dx.doi.org/10.1104/pp.004002
Lammers T., and Lavi S., 2007, Role of type 2C protein phosphatases in growth regulation and in cellular stress signaling, Crit Rev Biochem Mol Biol., 42(6): 437-461
Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., and Grill E., 2009, Regulators of PP2C phosphatase activity function as abscisic acid sensors, Science, 324(5930): 1064-1068
http://dx.doi.org/10.1126/science.1172408
Mustilli A., Merlot S., Vavasseur A., Fenzi F., and Giraudat J., 2002, Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell, 14(12): 3089-3099
http://dx.doi.org/10.1105/tpc.007906
Park S.Y., Fung P., Nishimura N., Jensen D.R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T.F., Alfred S.E., Bonetta D., Finkelstein R., Provart N.J., Desveaux D., Rodriguez P.L., McCourt P., Zhu J.K., Schroeder J.I., Volkman B.F., and Cutler S.R., 2009, Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins, Science, 324(5930): 1068-1071
http://dx.doi.org/10.1126/science.1172408
Schweighofer A., Hirt H., and Meskiene I., 2004, Plant PP2C phosphatases: emerging functions in stress signaling, Trends Plant, 9(5): 236-243
http://dx.doi.org/10.1016/j.tplants.2004.03.007
Seki M., Narusaka M., Kamiya A., Ishida J., Satou M., Sakurai T., Nakajima M., Enju A., Akiyama K., Oono Y., Muramatsu M., Hayashizaki Y., Kawai J., Carninci P., Itoh M., Ishii Y., Arakawa T., Shibata K., Shinagawa A., and Shinozaki K., 2002, Functional annotation of a full-length Arabidopsis cDNA collection, Science, 296(5565): 141-14
http://dx.doi.org/10.1126/science.1071006
Sheen J., 1998, Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants, Proc Natl Acad Sci. U S A., 95: 975-980
Tsugama D., Liu H., Liu S., and Takano T., 2012a, Arabidopsis heterotrimeric G protein β subunit interacts with a plasma membrane 2C-type protein phosphatase, PP2C52. Biochim Biophys Acta., 1823(12): 2254-2260
http://dx.doi.org/10.1016/j.bbamcr.2012.10.001
Tsugama D., Liu S., and Takano T., 2012b, A putative myristoylated 2C-type protein phosphatase, PP2C74, interacts with SnRK1 in Arabidopsis. FEBS Lett., 5866(6): 693-698
http://dx.doi.org/10.1016/j.febslet.2012.02.019
Umbrasaite J., Schweighofer A., Kazanaviciute V., Magyar Z., Ayatollahi Z., Unterwurzacher V., Choopayak C., Boniecka J., Murray J.A., Bogre L., Meskiene I., 2010, MAPK phosphatase AP2C3 induces ectopic proliferation of epidermal cells leading to stomata development in Arabidopsis, PLoS One, 5: e15357
http://dx.doi.org/10.1371/journal.pone.0015357
Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., Yamaguchi-Shinozaki K., Ishihama Y., Hirayama T., and Shinozaki K., 2009, Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci U S A. 106(41): 17588-17593
http://dx.doi.org/10.1073/pnas.0907095106
Wu F.H., Shen S.C., Lee L.Y., Lee S.H., Chan M.T., and Lin C.S., 2009, Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods. 24: 5-16
Xue T., Wang D., Zhang S., Ehlting J., Ni F., Jakab S., Zheng C.,and Zhong Y., 2008, Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics, 9: 550
http://dx.doi.org/10.1186/1471-2164-9-550
Yoshida R., Hobo T., Ichimura K., Mizoguchi T., Takahashi F., Aronso J., Ecker J.R., and Shinozaki K., 2002, ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis, Plant Cell Physiol, 43(12): 1473-1483
http://dx.doi.org/10.1093/pcp/pcf188
. PDF(318KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Hua Liu
. Daisuke Tsugama
. Shenkui Liu
. Tetsuo Takano
Related articles
. Protein phosphatase
. Vascular
. Protein-protein interaction
. Arabidopsis thaliana
Tools
. Email to a friend
. Post a comment


