2 Faculty of Science, Assuit University Alazher, Assuit, Egypt
 Author
Author  Correspondence author
Correspondence author
Genomics and Applied Biology, 2013, Vol. 4, No. 2 doi: 10.5376/gab.2013.04.0002
Received: 09 Jan., 2013 Accepted: 11 Feb., 2013 Published: 08 Apr., 2013
Hafez et al., 2013, Olive oil generated by the recombinant Escherichia coli with ACCase gene from Jatropha curcas, Genomics and Applied Biology, Vol.4 No.2 8-14 (doi: 10.3969/gab.2013.04.0002)
Acetyl-CoA carboxylase (ACCase) has a very important regulatory role in controlling plant fatty acid biosynthesis, thereby affecting lipid biosynthesis. The ACCase gene was amplified from Jatropha curcas using PCR with two degenerate primers, and the 1250-bp amplicon was cloned and sequenced. Sequence analysis revealed that the sequence obtained was similar to Jatropha curcas, Ricinus communis, Camellia sinensis and Phaseolus vulgaris acetyl cocoa, with 75% similarity. The full length of the gene was sub-cloned into a prokaryotic expression vector and the induced recombinant Escherichia coli was grown on 0.2% (w/v) sodium oleate. The cell metabolite was analyzed using thin-layer chromatography and HPLC. This analysis revealed that the cell metabolite consisted of a mixture of esters, mainly consisting of oleic acid (0.7 g/L) plus minor amounts of palmitic acid, linoleic acid and stearic acid. Fed-batch cultivation of E. coli was conducted at the specific growth rates of 0.15 and 0.1 h-1 for constant and exponential strategies, respectively. A high cell density of 20 g/L with an overall biomass yield of 3 g/L was achieved.
The use of biodiesel as a substitute fuel for the most of diesel engines is becoming attractive due to retreating petroleum assets and the environmental cost of exhaust gases from engines. Biodiesel, which means it is produced from many renewable sources mostly, consists of alkyl esters of fatty acids. It is considered as a potential prospect fuel, biodiesel has to be able to compete with petroleum diesel fuels, biodiesel has to be able to complete with petroleum diesel fuels on the economic level and on the environmental level as well. Veljkovic and his colleagues proposed that one way of tumbling biodiesel creation costs is less expensive compared with the by product of feedstocks contains fatty acids such as inedible oils, animal fats, and waste food oil (Veljkovic et al., 2006).
Vegetable oils of inedible plants produced by seed-forming trees and shrubs and can be substituted by Jatropha curcas which grows across the developing world (Openshaw, 2000). This tree Jatropha curca has no competing food uses. It can be used in Biodiesel production because their fatty acids have good characters more valuable when compared with the petroleum diesel (Krawczyk, 1996).
Estrification for the Plant oils must be carried out using the short-chain alcohols which could be used as a source of preferred green energy which should better for engine performances than fossil oil (Peterson C.L., 1995). Jatropha curca is probably the most highly promoted oilseed crop at present (Fairless, 2007). Much of the hype is reminiscent of that a few decades ago when many naive investors were separated from their capital by investing in plantations of jojoba, which was trumpeted as a crop that could grow in the desert without water and fertilizer and produce an oil that could replace sperm (whale) oil. In fact, both jojoba and Jatropha can grow in the desert without water and fertilizer, but not at commercial yields. High yields are only obtained after fertilizer and water inputs. The properties of Jatropha biodiesel was compared to those of diesel based on the American and European standards (Tiwarib, 2007).
The Enzyme acetyl-CoA carboxylase (ACCase, EC 6.4.1.2) play an important role in regulation of plant fatty acid biosynthesis. ACCase catalyzes the ATP-dependent carboxylation of acetyl-CoA to produce malonyl-CoA (Samols, 1988). The ACCase of eukaryotes (rather than plant) is comprised of multimers of a single polypeptide has multi-functional processes and it was isolated from different plant species (Kannangara, 1972). It is well known that the ACCase play an important role in several biochemical reactions and pathways in plants mainly in fatty acid synthesis and elongation (Harwood, 1988).
ACCase is multiple subcellular locations such as plastid and in the cell plasma membrane lipids (Cahoon, 1991). Fatty acids which synthesized outside the plastid considred as long chain fatty acids and their chains may contains more than 16 or 18-carbon atoms. Moreover, the malonyl- CoA which presented in the cell the cytosol is directed for fatty acids elongation reactions (Harwood, 1988). Malonyl-CoA is existing with different quantities according to the time and tissue (Post-Beittenmiller et al., 1993). In floral tissue, malonyl-CoA catalyze the chalcone synthase reaction which responsible to synthesize flavonoid pigments (Goodwin, 1983). In some tissues and under cretin circumstances such as the exposure of the UV lights, malonyl-CoA was transformed into in fatty acids which for membrane synthesis and maintenance (Ebel and Hahlbrock, 1977).
Many remarks have indicated that the ACCase is the sole enzyme responsible on oilseed fatty acid synthesis (Post-Beittenmiller et al., 1993). The main aim of this study was to produce biodiesel on a semi-industrial scale using recombinant E. coli. This work was the first attempt to produce oleic acid and increase the lipid contents in E. coli as a result of cloning the acetyl CoA carboxylase gene isolated from Jatropha plants. The lipids were extracted from recombinant E. coli grown in Erlenmeyer flasks and from E. coli grown on a semi-industrial scale using a biofermentor. The lipid contents in the cell were quantitatively and qualitatively analyzed by TLC and gas chromatography, and different extraction methods were investigated in order to maximize the yield in biodiesel.
Biodiesel considred as an interesting alternative energy source and to achievethis goal a novel approachs for establishing the biotechnology using recombinant organisms such as bacteria (Gu et al., 2011) different genes which encodes different subunits of the heteromeric ACCase were isolated, cloned and characterized Jatropha plant (accA, accB1, accC and accD). It well known that the ACCase also play a vital role in fatty acids biosynthesis in most of organism especially plants and algae (Gu et al., 2011). It was observed that there is a link between lipid deposition and ACCase enzyme activity in the mature seeds of oil crops (Deerberg et al., 1990). In mature seeds of the castor bean both of the ACCase activity and the expression levels of BC and BCCP were correlated with oil product (Roesler et al., 1996).
1 Results
The acetyl CoA enzyme was amplified from the Jatropha plant and fragments of about 1200 bp were obtained. Sequence alignment showed that a high level of similarity was observed when compared with the other acetyl CoA genes amplified from Jatropha, Ricinus, Camellia and Simarouba (Figure 1).

Figure 1 PCR amplification and comparative nucleotide DNA sequence analysis of the acetyl CoA gene
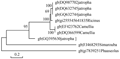
Phylogenetic construction of the acetyl CoA genes and the other eight CoA genes showed a high degree of similarity between the DNA sequence obtained and DQ632745 and DQ987702 (isolated from Jatropha), with an identity of 95%, similar to DQ632744 and FJ468293 with an identity of 99% (Jatropha and Simarouba, respectively) and 91% with 255545641 (Ricinus) (Figure 2; Figure3).
.png)
Figure 2 Phylogenetic tree showing the relationships between the nucleotide sequences of the acetyl CoA genes isolated from different plant genomes
The Camellia and Phaseolus genes showed a low degree of similarity with all the obtained genes. Jatropha curca is a tropical shrub that belongs to family Euphorbiaceous (Openshaw, 2000), Jatropha seeds high amount of oil ranged from 43~59% of the seed dry weight (Gubitz et al.,1999). Both of saturated and unsaturated fatty acids were founded in the jatropha with different concentrations and constituents (Augustus et al., 2002).
Accumulation of the lipid in the Acinetobacter baylyi strain ADP1 performed by the wax ester synthase/acyl-coenzyme A (Vaneechoutte et al., 2006). This enzyme able synthesizes wax esters and TAGs by catalyze either long-chain fatty acids or diacylglycerols (Kalscheuer et al., 2003). It was remaked that this enzyme has a broad substrate range including the short chain lengths and the very long chain ones (Kalscheuer et al., 2003; Kalscheuer et al., 2004; Stöveken et al., 2005; Uthoff et al., 2005).
The expression of WS/DGAT in recombinant hosts has shown that the promiscuity of this substrate has already been exploited to synthesize different molecules of fatty acid ester in vivo. The physiological background of the expression host regarding determined the type of fatty acid ester synthesized. Examples of these fatty acid ester derivatives are wax esters which produced in the recombinant Pseudomonas citronellolis (Kalscheuer et al., 2003). Additionally, fatty acid butyl esters (FABEs) in was produced by a recombinant Escherichia coli (Kalscheuer et al., 2006). Moreover, wax diesters and wax thioesters were produced by the mutant A. baylyi strain ADP1acr1 Km (Kalscheuer et al., 2003; Uthoff et al., 2005). Different types of fatty acids were produced by the recombinant yeast (In Saccharomyces cerevisiae) (Kalscheuer et al., 2004).
Fatty acid biosynthesis in E. coli was established by merging the expression of Jatropha curca acetyl CoA carboxylase genes using the pGEM-T and pPROEX™ HTa plasmids. The biosynthesis of FAEE was exactingly need to sodium oleate in the cultivation medium. The FAEEs that produced then gathered intracellularly, and no indication that an extracellular lipids were found in the culture filtrate. The HPLC and TLC analysis of FAEE isolated from E. coli TOP10 (pPROEX™ HTa) grown in medium complemented with sodium oleate, resulted in a mixture of esters that mainly consisted of oleic acid (0.7 g/L) plus minor amounts of palmitic acid, linoleic acid and stearic acid (Figure 4; Figure5).

Figure 4 HPLC analysis of oil produced from the recombinant E. coli
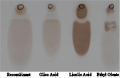
Figure 5 TLC analysis of the purified oil from the recombinant E. coli compared with the standard ones, Oleic acid, Linoleic acid and Ethyl oleate
Fed-batch fermentations were carried out in the previously mentioned medium (Gnansounou et al., 2005), but with addition of sodium oleate as a sole of carbon source. Over a similar time period, these fermentations revealed that the final titers of more than 3 g/L. Our results are supported by those of (Stephanopoulos et al., 2007). We can conclude that our recombinant E. coli was able to produce different fatty acids, particularly oleic acid, which could be a substitute for olive seed oil. In fed-batch experiments carried out under regulated aeration rates, (Kalscheuer et al., 2006) it was found that the highest FAEE levels were obtained. Optimized biodiesel manufacture by recombinant microorganisms could present some most important reward over reputable and predictable production processes.
Biotechnological biodiesel production could be considerably less expensive than usual biodiesel production if plant products such as starch or lignocellulose are used for its production. Plant oils are not only much expensive than plant polymers but plant oils are much more abundant, and they are the main source for biodiesel production regions of the world cultivated these plants. Biodiesel produced by microorganisms (recombinant) is considered as a completely sustainable biofuel that can be obtained from renewable resources with evade the use of methanol which is highly toxic.
2 Conclusion
In this study the team succeeded in producing a considerable amount of oil via a recombinant E. coli (transformed by the acetyl CoA synthase gene isolated from the Jatropha plant). The oil produced could substitute olive oil. Microbiodiesel is only one energy solution for the world right now. Because the discontinuous petroleum considered as a source of environmental pollution, whenever, the atomic energy is so dangerous and the world known very well what happened in Japan the last a few months.
In conclusion, this study provides a basis for achieving more competitive production costs and therefore a greater level of petroleum-derived fuel substitution by biofuels in the future.
3 Materials and Methods
3.1 Plant samples
The plant samples were obtained as shoot systems from tissue culture explants grown in jars, kindly provided by the Agriculture Research Center (ARC), Horticulture Research Institute, Giza, Egypt.
3.2 DNA extraction and PCR and acetyl CoA gene amplification
Jatropha plant leaves were subjected to DNA extraction using a Qiagen DNA extraction kit (Qiagen, Germany). The acetyl CoA carboxylase gene was amplified using primers we designed ourselves; forward (ME-75: 5' GTGGACCCATAGTTATGGCAACC 3') and reverse primer (ME- 76: 5' AGAAAGCTTCATCATTCCCCCAAG 3').
The PCR reaction was performed in a total volume of 25 μL, containing 2.5 μL10×buffer with MgCl, 2 μL 2.5 mmol/L dNTPs, 2 μL10 pmol of each primer, 2 μL DNA (100 ng/μL) and 0. 5 μL (5 units/μL) Taq DNA polymerase (Qiagen, Germany) and up to 25 μL dH2O. The PCR program consisted of: one cycle at 94℃ for 4 min, then 30 cycles consists of; denaturation step at 94℃ for 40s, annealing step at 52℃ for 40 s, and extension step at 72℃ for 1.5 min. The reaction was ended by one cycle at 72℃ for 10 min. Finally, the reaction was stored at 4 ℃ until be used.
3.3 Polymorphism and divergence
Sequence alignment were carried out using CLUSTALW (1.82) (Thompson et al., 1994). A bootstrap neighbour-joining tree was constructed using the program MEGA 2.1 (Kumar et al., 2001) from CLUSTALW alignments.
3.4 DNA Purification and cloning in the TOPO TA cloning vector
The PCR amplicone was visulaized on 1% low-melting-point agarose gel and the DNA band was cut from the gel using a sterile, sharp scalpel and the agarose-embedded DNA fragments were then transferred to a sterile 2 mL Eppendorf tube. The DNA purification was performed using an agrose DNA extraction kit (Qiagen, Germany). The PCR product was eluted in 30 µL H2O, and 2 µL of the purified PCR product was transferred into the pCR 2.1 TOPO vector (TOPO TA cloning kit, Invitrogen). Recombinant E. coli cells were subjected to plasmid DNA extraction using a mini-plasmid DNA kit (Qiagen). The full-length Jatropha ACCase was released from the pCR® 2.1-TOPO® vector using an EcoRI restriction enzyme; meanwhile, the released fragment (1.2 kbp) was purified using an EzWayTMGel Extraction kit (KOMBIOTECH, Korea) and ligated to the linearized prokaryotic expression pPROEX HT (Life Technologies, USA), and the transformation reaction was performed according to the protocols outlined by Life Technologies (Invitrogen).
3.5 Bioreactor and feed-batch
Fed-batch cultivations were performed in a 3 L bench-top bioreactor (Bioflow III, New Brunswick, NJ, USA) equipped with two 6-bladed disc-turbine impellers and four baffles and connected to a digital control unit. The process was automated through a computer-aided data bioprocessing system, the AFS-BioCommand multi-process management program. The set points for temperature 30℃ and pH 7, respectively, and the pH level was controlled by the automatic feeding of 2 N NaOH. Compressed air was initially supplied at 0.5-1.0 VVM (air volume per broth volume per min) through a sterile filter. This could be manually controlled in parallel with the agitation speed (150~900 rpm) to maintain the dissolved oxygen level above 20%. The dissolved oxygen level and the pH values were controlled and measured on-line with METTLER TOLEDO electrodes. Antifoam A (Sigma) was used to eliminate foaming. A 0.2% (w/v) sodium oleate, 2% (w/v) glucose, 1 mmol/L IPTG and appropriate antibiotics was added to the LB medium. The medium was inoculated with 5% (v/v) of our inoclum.
3.6 Thin-layer chromatography
The TLC analysis was performed for the total lipid extracted from the recombinant bacterium (E. coli) according to described (Kalscheuer and Steinbüchel 2003). Lipids were separating by spraying with 40% (v/v) sulphuric acid and charring. Oleic acid, linoleic acid and ethyl oleate were purchase from Sigma-Aldrich and used as reference substances for the FAEEs.
3.7 HPLC analysis of fatty acids
Fatty acids in the methanol extracts from the oils were then analyzed by high-performance liquid chromatography (HPLC) (Agilent 1100, USA) using a Capcell Pak C18 column (25 cm in length, 4.6 mm in inner diameter, 5 lm) and an ltraviolet detector at 220 nm (Tosoh, UV-8024) operated at 30℃ with 0. 7 mL/min flow rate of 80% acetonitrile solution containing 20% of 0.1% H3PO4 as a carrier solvent. The sample volume was 20 μL and the peaks were identified by comparing the reaction times between the sample and the standard compound.
References
Augustus, G.S., Jayabalan M., and Seiler G.J., 2002, Evaluation and bioinduction of energy components of Jatropha curcas, Biomass and Bioenergy, 23(3): 161-164
http://dx.doi.org/10.1016/S0961-9534(02)00044-2
Cahoon, D.R., Levine J.S., Cofer III W.R., Minnis P., Miller J.E., Tennile G.M., Yip T.W., Heck P.W., and Stocks B.J., 1991, The Great Chinese Fire of 1987: a view from space. In: Global Biomass Burning: Atmospheric, Climatic, and Biospheric Implications (J.S.Levine, ed.), MIT Press, Cambridge, MA, pp. 61-66
Deerberg S., Wickel J.V., Förster H.H., Cole T., Fuhrmann J., and Heise K.P., 1990, Synthesis of medium-chain fatty acids and their incorporation into triacylglycerols by cell-free fractions from Cuphea embryos, Planta, 180(3): 440-444
Ebel J., and Hahlbrock K., 1977, Enzymes of flavone and flavonol glycoside biosynthesis. Coordinated and selective induction in cellsuspension cultures of Petroselinum hortense, Eur. J Biochem., 75(1), 201-209
http://dx.doi.org/10.1111/j.1432-1033.1977.tb11518.x
Fairless D., 2007, Biofuel: the little shrub that could—maybe, Nature, 449: 652–655
http://dx.doi.org/10.1038/449652a
Gnansounou E., Dauriat A., and Wyman C.E., 2005, Refining sweet sorghum to ethanol and sugar: economic trade-offs in the context of North China, Bioresource Technology, 96(9): 985-1002
Goodwin N.S., Park P.J.D., and Rawlinson A.P., 1983, Crude oil biodegradation under simulated and natural condition, In M. Bjorøy, and et al., eds., Advances in Organic Geochemistry 1981, J. Wiley & Sons, New York, pp.650-658
Gu K., Chiam H., Tian D., and Yin Z., 2011, Molecular cloning and expression of heteromeric ACCase subunit genes from Jatropha curcas, Plant Sci., 180(4), 642-649
Gubitz G.M., Mittelbach M., and Trabi M., 1999, Exploitation of the tropical oil seed plant Jatropha curcas L., Bioresour. Technol., 67(1): 73–82
http://dx.doi.org/10.1016/S0960-8524(99)00069-3
Harwood J.L., 1988, Fatty acid metabolism, Annu. Rev. Plant Physiol. Plant MoI. Biol. 39, 101–138
Kalscheuer R. and Steinbüchel A., 2003, A novel bifunctional wax ester synthase/acyl-CoA: diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1, J Biol. Chem., 278(10): 8075-8082
Kalscheuer R., Luftmann H., and Steinbüchel A., 2004, Synthesis of novel lipids in Saccharomyces cerevisiae by heterologous expression of an unspecific bacterial acyltransferase, Appl. Environ. Microbiol., 70(12): 7119–7125
Kalscheuer R., Stöveken T., Luftmann H., Malkus U., Reichelt R., and Steinbüchel1 A., 2006, Neutral lipid biosynthesis in engineered Escherichia coli: jojoba oil-like wax esters and fatty acid butyl esters, Appl. Environ. Microbiol., 72(2): 1373–1379
http://dx.doi.org/10.1128/AEM.72.2.1373-1379.2006
Kannangara C.G., and Stumpf P.K., 1972, Fat metabolism in higher plants ☆: L. The biosynthesis of polyunsaturated fatty acids by isolated spinach chloroplasts, Arch. Biochem. Biophys, 148(2): 414-424
http://dx.doi.org/10.1016/0003-9861(72)90159-2
Krawczyk T., 1996, Biodiesel alternative fuel makes inroads but hurdles remain, INFORM, 7: 801–829
Kumar J.P., Wilkie G.S., Tekotte H., Moses K., and Davis I., 2001, Perturbing nuclear transport in Drosophila eye imaginal discs causes specific cell adhesion and axon guidance defects, Dev. Biol., 240(2): 315-325
Openshaw K., 2000, A review of Jatropha curcas: An oil plant of unfulfilled promise. Biomass and Bioenergy, 19(1): 1–15
http://dx.doi.org/10.1016/S0961-9534(00)00019-2
Peterson C.L., Hammond B., Reece D., Thompson J., and Beck S., 1995, Performance and durability testing of diesel engines using ethyl and methyl ester fuels. Report submitted in completion for contracts 236-l and 52016-l from the National Biodiesel Board USA. Department of Biological and Agricultural Engineering, University of Idaho, Moscow, USA.
Post-Beittenmiller D., Ohlrogge J.B., and Somerville C.R., 1993, Regulation of Plant Lipid Biosynthesis: An Example of Developmental Regulation Superimposed on a Ubiquitous Pathway, In: Verma D.P.S., ed., Control of Plant Gene Expression, CRC press, Boca Raton, Fia., pp.157-174
Roesler K.R.,Savage L.J., Shintani D.K., Shorrosh B.S., and Ohlrogge J.B., 1996, Co-purification, co-immunoprecipitation, and coordinate expression of acetyl-coenzymeAcarboxylase activity, biotin carboxylase, and biotin carboxyl carrier protein of higher plants, Planta, 198(4): 517–525
Saitou N., and Nei M., 1987, The neighbor-joining method: a new method for reconstructing phylogenetic trees, Mol Biol Evol., 4(4): 406-425
Samols D., Thornton C.G., Murtif V.L., Kumar G.K., Haase F.C., and Wood H.G., 1988, Evolutionary conservation among biotin enzymes, J. Biol. Chem., 263(14): 6461-6464
Stephanopoulos G., 2007, Challenges in engineering microbes for biofuels production. Science, 315 (5813): 801-804
http://dx.doi.org/10.1126/science.1139612
Stöveken, T., Kalscheuer, R., Malkus, U., Reichelt, R. and Steinbüchel, A., 2005, The wax ester synthase/acyl coenzyme A: diacylglycerol acyltransferase from Acinetobacter sp. strain ADP1: characterization of a novel type of acyltransferase, J Bacteriol., 187(4): 1369-1376
http://dx.doi.org/10.1128/JB.187.4.1369-1376.2005
Thompson J.D., Higgins D.G., and Gibson T.J., 1994, CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice, Nuc. Ac. Res., 22(22): 4673-4680
Tiwarib A.K., Kumar A., and Raheman H., 2007, Biodiesel production from jatropha oil (Jatropha curcas) with high free fatty acids: An optimized process, Biomass Bioenergy, 31(8): 569–575
http://dx.doi.org/10.1016/j.biombioe.2007.03.003
Uthoff S., Stoveken T., Weber N., Vosmann K., Klein E., Kalscheuer R., and Steinbuchel A., 2005, Thio wax ester biosynthesis utilizing the unspecific bifunctional wax ester synthase/acyl-CoA : diacylglycerol acyltransferase of Acinetobacter sp. strain ADP1, Appl. Environ. Microbiol., 71(2): 790-796
http://dx.doi.org/10.1128/AEM.71.2.790-796.2005
Vaneechoutte, M., Young D. M., Ornston L. N., De Baere T., Nemec A., Van Der Reijden T., Carr E., Tjernberg I., and Dijkshoorn L., 2006, Naturally transformable Acinetobacter sp. strain ADP1 belongs to the newly described species Acinetobacter baylyi, Appl Environ. Microbiol., 72(1): 932-936
http://dx.doi.org/10.1128/AEM.72.1.932-936.2006
Veljkovic V.B., Lakicevic S.H., Stamenkovic O.S., Todorovic Z.B. and Lazic M.L., 2006, Biodiesel production from tobacco (Nicotiana tabacum L.) seed oil with a high content of free fatty acids, Fuel, 85(17-18): 2671–2675
http://dx.doi.org/10.1016/j.fuel.2006.04.015
. PDF(420KB)
. HTML
Associated material
. Readers' comments
Other articles by authors
. Elsayed E. Hafez
. Usama M
. Ubdul -Rahouf
. Salahedin G. Ali
. Elsayed K. Bakhiet
Related articles
. Acetyl Co-A carboxylase gene
. Jatropha curcas
. Biodiesel
. Oleic acid
. Palmitic acid
. Linoleic acid
. Stearic acid
Tools
. Email to a friend
. Post a comment