Cloning and Expression Analysis of β-ketoacyl-ACP Reductase, β-hydroxyacyl-ACP Dehydrase and Enoyl-ACP Reductase from Arachis hypogaea L. 
 Author
Author  Correspondence author
Correspondence author
Genomics and Applied Biology, 2010, Vol. 1, No. 2 doi: 10.5376/gab.2010.01.0002
Received: 15 Aug., 2010 Accepted: 19 Sep., 2010 Published: 28 Sep., 2010
This is an Open Access article published under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Chi et al., 2010, Cloning and Expression Analysis of β-ketoacyl-ACP Reductase, β-hydroxyacyl-ACP Dehydrase and enoyl-ACP Reductase from Arachis hypogaea L., Genomics and Applied Biology, 2010, Vol. 1, No. 2 (doi: 10.5376/gab.2010.01.0002)
Fatty acid biosynthesis is catalysed in most bacteria and plants by a group of highly conserved proteins known as the Type II fatty acid synthase (FAS) system. In this study, genes for β-ketoacyl-ACP reductase (KR), β-hydroxyacyl-ACP dehydrase (HD), and enoyl-ACP reductase (ENR) of Type II FAS have been cloned from peanut (Arachis hypogaea L.). The results showed that the ORF of the three genes were 972 bp, 651 bp and 1170 bp in length, encoding 323, 216 and 389 amino acids, respectively. The predicted amino acid sequences of AhKR, AhHD, AhENR shared high sequence identity of 86.3%, 81.2% and 87.2% to the corresponding ones in Glycine max, respectively. Further investigation of quantitative real-time RT-PCR analysis suggested that AhKR, AhHD and AhENR were expressed with higher levels in leaf and seed than those in root and stem tissues. Moreover, AhKR and AhENR genes reached a maximum expression level at 25 DAP (days after pegging) and showed a downward trend thereafter. In contrast, AhHD gene expressed in an irregular course during seed development. Overall, the information generated by this study will facilitate the manipulation of the quality of oils produced in seeds of oil crops.
Fatty acid synthesis is essential for the formation of membranes and hence for the viability of all organisms (Massengo-Tiassé and Cronan, 2007). A complex enzyme system, fatty acid synthetase (FAS), is used throughout nature for the de novo biosynthesis of fatty acids from acetyl-CoA and malonyl-CoA (Fisher et al., 2000). In general, FAS is organized into two different systems (FAS I and FAS II) based on the architecture of the enzymes involved. In FAS I system, the involved synthases found in fungi and mammals are large multifunctional enzymes with multiple domains that catalyze various reactions of FAS (Smith et al., 2003; Schweizer and Hofmann, 2004). In contrast, the FAS of bacteria, chloroplasts, apicoplasts and mitochondria belongs to the FAS II system, where the acyl chain covalently attached to the acyl carrier protein (ACP) is elongated with four enzymes catalyzing consecutively (Campbell and Cronan, 2001; Olsen et al., 2004).
In bacteria and plants, the enzymes which catalyze the four successive steps of the pathway are as follows: β-ketoacyl-acyl carrier protein (ACP) synthetases I, II and III; β-ketoacyl-acyl carrier protein (ACP) reductase; β-hydroxyacyl-acyl carrier protein (ACP) dehydratase; and enoyl-acyl carrier protein (ACP) reductase (Rafferty et al., 1995). Chain termination is mediated by the hydrolysis of acyl-ACP, catalyzed by an acyl-ACP thioesterase. The fatty acyl chain lengthens by two carbons each time it cycles through the enzymatic reactions (Tai et al., 2007). Most fatty acids reach a chain length of 16 or 18 carbons and then are targeted for glycerolipid synthesis in the plastid or in the endoplasmic reticulum (Browse and Somerville, 1991).
β-ketoacyl-ACP reductase (KR) catalyzes the pyridine-nucleotide-dependent reduction of a β-oxoacyl form of acyl carrier protein (ACP), the second step in de novo fatty acid biosynthesis and a reaction often performed in polyketide biosynthesis (Fisher et al., 2000). Two isoforms of the enzyme, one NADPH-linked and the other NADH-linked, have been reported (Caughey and Kekwick, 1982). The function of the NADH-linked enzyme is unknown. It is the NADPH-linked β-ketoacyl-ACP reductase that functions in fatty acid biosynthesis (Slabas et al., 1992). KR is a member of the Short-chain Dehydrogenase/Reductase (SDR) superfamily (Price et al., 2001), and thereby displays the amino acid signature of this family, Ser-X12-Tyr-X3-Lys (Persson et al., 2003).
The third step in the elongation cycle is catalyzed by β-hydroxyacyl-ACP dehydratase (HD). There are two isoforms. FabZ, which catalyzes the dehydration of (3R)-hydroxyacyl-ACP to trans-2-acyl-ACP, is a universally expressed component of the type II system. FabA, the second isoform, as has more limited distribution in nature and, in addition to dehydration, also carries out the isomerization of trans-2-to cis-3-decenoyl-ACP as an essential step in unsaturated fatty acid biosynthesis (Heath and Rock, 1996; Kimber et al., 2004). The catalytic site is hydrophobic except for a histidine and a glutamine (aspartic acid), which together are proposed to catalyze the reactions (Leesong et al., 1996).
The enoyl-ACP reductase (ENR) catalyses the last step in the fatty acid elongation cycle (Marrakchi et al., 2003). Four types of ENR, namely, FabI, fabK, FabL and FabV, have been reported (Bergler et al., 1994;Heath et al., 2000; Marrakchi et al., 2003; Massengo-Tiassé and Cronan, 2007). Most ENRs (FabI, FabL and FabV) are distant members of the SDR superfamily. Other classes of ENRs are the TIM barrel flavin containing enzyme, FabK, the ENRs found in mitochondria and the ENR domains of the mammalian and fungal megasynthases. Both NADH and NADPH are used as the hydride source in the reactions of SDR-type ENRs (Massengo-Tiassé and Cronan, 2007). FabI and FabL are atypical in that the key residues are a diad consisting of a Tyr-X6-Lys motif in contrast with the Tyr-X3-Lys in prototypical SDRs (Baker, 1995; Parikh et al., 1999). FabV is considerably larger than either FabI or FabL, and contains a Tyr-X8-Lys active site motif (Massengo-Tiassé and Cronan, 2007).
Cultivated peanuts (Arachis hypogaea L.) are important oilseed crops worldwide, because these allotetraploids (2n=4x=40) typically contain 50% oil in the seed. The flavor and quality of either the seed or the oil is immensely dependent on the fatty acid composition of the extracted oil (Andersen and Gorbet, 2002; Jung et al., 2000b; Lopez et al., 2000; Yu et al., 2008). It would be of great importance to study the fatty acid biosynthesis pathway for improving oil quality and increasing oil content of peanut. In this study, we isolated and characterized cDNAs containing the complete coding region of AhKR, AhHD and AhENR genes, and analyzed their expression in different organs and at different development stages of seeds. The identification of these novel genes in this study will be helpful for the reconstruction of the pathways involved in fatty acid biosynthesis and the metabolic engineering of fatty acid synthesis in peanut.
1 Results
1.1 Isolation and analysis of AhKR, AhHD and AhENR genes
Three full-length genes namely β-ketoacyl-ACP reductase (KR), β-hydroxyacyl- ACP dehydrase (HD), and enoyl-ACP reductase (ENR) were isolated from a peanut seedling full-length cDNA library (unpublished data). The ORF of the three genes were 972 bp, 651 bp and 1170bp in length, encoding 323, 216 and 389 amino acids, respectively (Table 1). Prediction of subcellular location by two programs, TargetP Server and Predotar, suggested that these three proteins probably located in chloroplast. The first 73, 43 or 70 amino acids at the N-terminal end of the deduced protein for AhKR, AhHD or AhENR had a high proportion of hydroxylated and small, hydrophobic amino acids, which was typical of chloroplast transit peptide. A Blast search revealed that the primary structure of AhKR, AhHD, AhENR shared high sequence identity of 86.3%, 81.2% and 87.2% to the corresponding ones in Glycine max, respectively. The deduced amino acid sequences of AhKR, AhHD, AhENR showed 37.3%, 30.2%, and 20.9% identity with EcFabG, EcFabZ, and EcFabI, respectively.
![]()
Table 1 AhKR, AhHD and AhENR genes in peanut
1.1.1 Cloning and phylogenetic analysis of AhKR gene
In Figure 1, the peanut β-ketoacyl-ACP reductase protein sequence was compared for similarity with those of plants and bacteria using ClustalW. A triad of Ser217, Tyr230 and Lys234 residues involved in catalysis and substrate binding was revealed by sequence alignments, characteristic of the SDR family. The tyrosine and lysine residues are involved in actual catalysis, whereas serine participates in substrate binding and alignment (Price et al., 2001; Li et al., 2009). To examine the relationships among different sources of KR genes, the neighbour-joining method was used to construct the phylogenetic tree (Figure 2). As shown in the phylogenetic tree, all of the KR genes fell into two subfamilies: the bacteria subfamily and the cyanobacteria/green algae/mosses/higher plants subfamily. The AhKR gene from peanut clustered with those from higher plants, and the genes from cyanobacteria may be the origin of genes from higher plants, mosses and eukaryotic algae.
 Figure 1 Multiple sequence alignments of KR homologs |
 Figure 2 Neighbor-joining tree based on the deduced amino acid sequences of KR homologs |
1.1.2 Cloning and phylogenetic analysis of AhHD gene
As shown in Figure 3A, multiple sequence alignment indicated that AhHD protein contained a histidine at amino acid 116 and a glutamine at amino acid 130, which were in positions that were conserved in the FabZ of Escherichia coli (Cronan et al., 1988). The location of these residues corresponded to the catalytically active site of the enzymes in Plasmodium faliciparum and Escherichia coli. (Cronan et al., 1988; Heath and Rock, 1996; Sharma et al., 2003). Specifically, there were distinct differences in the active site residues, an Asp in FabA and a Glu in FabZ (Figure 3B). In addition, similar to PaFabZ (Kimber et al., 2004), ï¬ve key motifs, LPHRFPFLLVD, GHFP, PGVL, EAMAQ, and AGD, were also conserved in AhHD. Phylogenetic analysis suggested that fabZ and fabA genes fell into two separate subfamilies (Figure 4). The AhHD gene from peanut clustered together with genes from higher plants, and were separated from those of green algae. The FabZ genes from cyanobacteria may be the origin of genes from higher plants and eukaryotic algae.
.png) Figure 3 Multiple sequence alignments of HD homologs |
.png) Figure 4 Neighbor-joining tree based on the deduced amino acid sequences of HD homologs |
1.1.3 Cloning and phylogenetic analysis of AhENR gene
FabV, FabI and FabL were distant members of the SDR superfamily, although FabV aligned only weakly with FabI or FabL even when many gaps were allowed (Figure 5A). FabI and FabL contained a Tyr-X6-Lys active site motif. In FabV other variation was seen, a Tyr-X8-Lys motif (Figure 5A). The spacing between the putative FabV active site tyrosine and lysine residues was eight residues, two more than FabI and FabL and one more than the maximum reported for other SDR proteins. FabK, first discovered in Streptococcus pneumoniae, was refractory to triclosan and was a flavoprotein unrelated to the SDR isozymes (Marrakchi et al., 2003). The alignment of the predicted FabK-like proteins from this subset of organisms demonstrated that they possessed a conserved nucleotide-binding domain in the N-terminus and a conserved flavin-binding domain at the centre of the protein (Figure 5B).
.png) Figure 5 Multiple sequence alignments of ENR homologs |
AhENR and AhKR were members of the SDR superfamily, and AhENR showed 16.1% sequence identity with AhKR, compared with 25% in Brassica napus (Fisher et al., 2000). However, KR and ENR catalyzed distinctly different chemical reactions, namely a carbon–oxygen double-bond reduction and a carbon-carbon double-bond reduction, respectively. Fisher et al. (2000) reported that striking similarities existed in fold, mechanism and substrate binding of BnKR and BnENR. Thus, these two enzymes may have diverged from a common ancestor during the evolution of the biosynthetic pathway. Phylogenetic analysis suggested that the AhENR gene from peanut formed a group with the genes from higher plants and green algae, and set apart from the groups of cyanobacteria and bacteria (Figure 6).
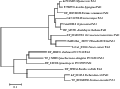 Figure 6 Neighbor-joining tree based on the deduced amino acid sequences of ENR homologs |
The quantitative real-time RT-PCR (qRT-PCR) was employed to confirm the expression patterns of AhKR, AhHD, and AhENR genes in four peanut tissues and at different developmental stages of seeds. β-actin was used as an internal reference control for total RNA input. β-actin PCR product was not detected when reverse transcriptase was omitted, indicating that the RNA template was free of genomic DNA. The results revealed that the three genes were expressed dominantly in leaf among four tissues tested, and expressed at the lowest level in stem (Figure 7). In addition, the expression level of these genes in seed was also relatively high. The expression patterns of the three genes across six developmental stages of seed were illustrated in Figure 8. AhKR and AhENR genes reached a maximum expression level at 25 DAP and showed a downward trend thereafter. In contrast, AhHD gene expressed in an irregular course during seed development and had high expression at 25 and 60 DAPs. These results indicated that these genes may have different biochemical functions during vegetative growth and seed development.
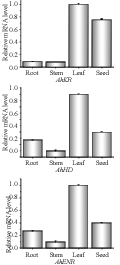 Figure 7 Expression analysis of AhKR, AhHD and AhENR genes in four different tissues |
 Figure 8 Expression analysis of AhKR, AhHD and AhENR genes in seed at different developmental stagesAX |
2 Discussion
Peanut is an important crop internationally for both food and oil production (Luo et al., 2005). One of the major factors influencing peanut oil quality is the composition of fatty acid. So validating the mechanism of the fatty acid synthesis and metabolism is the central goal to increase peanut quality. Fatty acid biosynthesis in higher plants is carried out by type II FAS. In the present study, we isolated AhKR, AhHD and AhENR orthologues in peanut seedling. Sequence comparisons revealed that the primary structure of plant plastid type II FAS enzymes (KR, HD, ENR) was strictly conserved, especially the catalytic residues, which suggested that these enzymes may have similar functions in higher plants as those in E. coli. Real-time RT-PCR analysis revealed that the three genes were expressed dominantly in leaf among four tissues tested, and expressed at the lowest level in stem. AhKR and AhENR genes shared similar expression behaviors over the developmental stages compared to that of AhHD gene. These results indicated that the three genes may have different biochemical functions during vegetative growth and seed development. To improve peanut oil quality should coordinate the expression of the three enzymes involved in the fatty acid synthesis pathway. This work may provide a basis for elucidating the molecular mechanism of fatty acid synthesis and provide candidate genes for modifying oil quality via transgenic plants.
3 Materials and methods
3.1 Plant materials
Peanut seeds (Arachis hypogaea L. cultivar Huayu19) were sown in sand and soil mixture (1:1), grown in a growth chamber under a 16 h~8 h light-dark cycle at 26°C and 22°C, respectively. Three kinds of 12-day-old tissues including root, stem and leaf were collected as experimental materials for quantitative real-time RT-PCR analysis. In addition, the immature peanut seeds from 25 to 60 days after pegging (DAP) were also collected for expression analysis.
3.2 Nucleic acid manipulation
Total RNA was extracted from samples using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. The RNA samples were used for real-time RT-PCR after RQ1 RNase-free DNaseI (Promega, Wisconsin, USA) treatment to remove genomic DNA. The ï¬rst-strand cDNA was synthesized with RT-PCR kit (Promega, Wisconsin, USA) using 500 ng of total RNA according to the manufacturer’s instructions. Controls received water instead of reverse transcriptase to assess any contamination from genomic DNA as described by Zhou et al. (2007).
3.3 Full-length cDNA sequence isolation
PCR was performed with the LA PCR system (Takara) using 2.5 μl of 10×PCR buffer with MgCl2, 1 μl of 10 μM each primer (Table 2), 4.0 μl of 10 mM dNTPs, 1 μl cDNA samples and 0.5 μl LA Taq™ DNA polymerase, and 15 μl double distilled water. The PCR products were run on 1% agarose gel and purified with Gel Extraction Kit (Takara) according to the manufacturer’s protocol. The purified products were then cloned into the pMD18-T Easy vector (Takara) and sequenced (Shangon, Shanghai).
 Table 2 Primers used in experiment |
3.4 Multiple sequence alignment and phylogenetic analysis
Physicochemical properties of the deduced protein were predicted by Protparam (http://www.expasy.ch/tools/ protparam.html). The putative subcellular localizations of the candidate proteins were estimated by TargetP (http://www. cbs.dtu.dk/services/TargetP/) and Predotar (http://urgi.versailles.inra.fr/predotar/predotar.html). The potential N-terminal presequence cleavage site was predicted by ChloroP (http://www.cbs.dtu.dk/services/ChloroP/). Amino acid sequences were aligned using ClustalX program with the implanted BioEdit (Thompson et al., 1994). The neighbor-joining (NJ) method in MEGA4 (Tamura et al., 2007) was used to construct the phylogenetic tree. Bootstrap with 1000 replicates was used to establish the confidence limit of the tree branches. Default program parameters were used.
3.5 Quantitative real-time RT-PCR
The real-time RT-PCR analysis was performed by using a LightCycler 2.0 instrument system (Roche, Germany). β-actin gene was taken as reference gene. Three pairs of gene-speciï¬c primers (Table 1) were designed according to the AhKR, AhHD and AhENR gene sequences. The real-time RT-PCR reactions were performed by using the SYBR Premix Ex Taq polymerase (TaKaRa, Japan) according to the manufacturer’s instructions. The expression of the gene was calculated relative to the calibration sample and the β-actin to normalize the sample input amount. All the experiments were performed in triplicate to ensure the data accuracy.
References
Andersen P.C., and Gorbet D.W., 2002, Influence of year and planting date on fatty acid chemistry of high oleic acid and normal peanut genotypes, J Agric Food Chem., 50: 1298-1305 doi:10.1021/jf0113171 PMid:11853521
Baker M.E., 1995, Enoyl-acyl-carrier-protein reductase and Mycobacterium tuberculosis InhA do not conserve the Tyr-Xaa-Xaa-Xaa-Lys motif in mammalian 11b-and 17b-hydroxysteroid dehydrogenases and Drosophila alcohol dehydrogenase, Biochem. J., 309: 1029-1030 PMid:7639680 PMCid:1135734
Bergler H., Wallner P., Ebeling A., Leitinger B., Fuchsbichler S., Aschauer H., Kollenz G., Hogenauer G., and Turnowsky F., 1994, Protein envM is the NADH-dependent enoyl-ACP reductase fabI of Escherichia coli, J. Biol. Chem., 269: 5493-5496 PMid:8119879
Browse J., and Somerville C., 1991, Glycerolipid synthesis: biochemistry and regulation, Ann. Rev. Plant Physiol. Plant Mol. Biol., 42: 467-506 doi:10.1146/annurev.pp.42.060191.002343
Campbell J.W., and Cronan Jr J.E., 2001, Bacterial fatty acid biosynthesis: Targets for antibacterial drug discovery, Annu. Rev. Microbiol., 55: 305-332 doi:10.1146/annurev.micro.55.1.305 PMid:11544358
Caughey I., and Kekwick G.O., 1982, The characteristics of some components of the fatty acid synthetase system in the plastids from the mesocarp of avocado (Persea americana) fruit, Eur. J. Biochem., 123: 553-561 doi:10.1111/j.1432-1033.1982.tb06568.x
Cronan Jr J.E., Li W.B., Coleman R., Narasimhan M., de Mendoza D., and Schwab J.M., 1988, Derived amino acid sequence and identiï¬cation of active site residues of Escherichia coli beta-hydroxydecanoyl thioester dehydrase, J. Bio. Chem., 263: 4641-4646
Fisher M., Kroon J.T., Martindale W., Stuitje A.R., Slabas A.R., and Rafferty J.B., 2000, The X-ray structure of Brassica napus beta-keto acyl carrier protein reductase and its implications for substrate binding and catalysis, Structure, 8(4): 339-347 doi:10.1016/S0969-2126(00)00115-5
Heath R., and Rock C., 1996, Roles of the FabA and FabZ beta-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis, J. Biol. Chem., 271: 27795-2780 doi:10.1074/jbc.271.44.27795 PMid:8910376
Heath R.J., Su N., Murphy C.K., and Rock C.O., 2000, The Enoyl-[acyl-carrier-protein] Reductases FabI and FabL from Bacillus subtilis, J. Biol. Chem., 275: 40128-40133 doi:10.1074/jbc.M005611200 PMid:11007778
Heath, R.J. and Rock C.O., 1995, Enoyl-acyl carrier protein reductase (fabI) plays a determinant role in completing cycles of fatty acid elongation in Escherichia coli, J. Biol. Chem., 270: 26538-26542 doi:10.1074/jbc.270.44.26538 PMid:7592873
Jung S., Swift D., Sengoku E., Patel M., Teule F., Powell G., Moore K., and Abbott A., 2000, The high oleate trait in the cultivated peanut [Arachis hypogaea L.]. I. Isolation and characterization of two genes encoding microsomal oleoyl-PC desaturases, Mol. Gen. Genet., 263: 796-805 doi:10.1007/s004380000244
Kimber M.S., Martin F., Lu Y., Houston S., Vedadi M., Dharamsi A., Fiebig K.M., Schmid M., and Rock C.O., 2004, The structure of (3R)-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Pseudomonas aeruginosa, The Journal of Biological Chemistry, 279(50): 52593-52602 doi:10.1074/jbc.M408105200 PMid:15371447
Leesong M., Henderson B.S., Gillig J.R., Schwab J.M., and Smith J.L., 1996, Structure of a dehydratase–isomerase from the bacterial pathway for biosynthesis of unsaturated fatty acids: two catalytic activities in one active site, Structure, 4(3): 253-264 doi:10.1016/S0969-2126(96)00030-5
Li M.J., Li A.Q., Xia H., Zhao C.J., Li C.S., Wan S.B., Bi Y.P., and Wang X.J., 2009, Cloning and sequence analysis of putative type II fatty acid synthase genes from Arachis hypogaea L, J. Biosci., 34(2): 227-238 doi:10.1007/s12038-009-0027-1 PMid:19550039
Lopez Y., Nadaf H.L., Smith O.D., Connell J.P., Reddy A.S., and Fritz A.K., 2000, Isolation and characterization of the Delta(12)-fatty acid desaturase in peanut (Arachis hypogaea L.) and search for polymorphisms for the high oleate trait in Spanish market-type lines, Theor. Appl. Genet., 101: 1131-1138 doi:10.1007/s001220051589
Luo M., Dang P., Guo B.Z., He G., Holbrook C.C., Bausher M.G., and Lee R.D., 2005, Generation of expressed sequence tags (ESTs) for gene discovery and marker development in cultivated peanut, Crop Sci., 45: 346-353 doi:10.2135/cropsci2005.0346
Marrakchi H., Dewolf Jr W.E., Quinnœ C., West J., Polizzi B.J., So C.Y., Holmes D.J., Reed S.L., Heath R.J., Payne D.J., Rock C.O., and Wallis N.G., 2003, Characterization of Streptococcus pneumoniae enoyl-(acyl-carrier protein) reductase (FabK), Biochem. J., 370: 1055-1062 doi:10.1042/BJ20021699 PMid:12487627 PMCid:1223239
Massengo-Tiassé R.P., and Cronan J.E., 2007, Vibrio cholerae fabV defines a new class of enoyl acyl-carrier-protein reductase, The Journal of Biological Chemistry, 283(3): 1308-1316 doi:10.1074/jbc.M708171200 PMid:18032386
Massengo-Tiassé R.P., and Cronan J.E., 2009, Diversity in enoyl-acyl carrier protein reductases, Cell Mol. Life Sci., 66(9): 1507-1517 doi:10.1007/s00018-009-8704-7 PMid:19151923 PMCid:2819910
Olsen J.G., Rasmussen A.V., von Wettstein-Knowles P., and Henriksen A., 2004, Structure of the mitochondrial beta-ketoacyl-[acyl carrier protein] synthase from Arabidopsis and its role in fatty acid synthesis, FEBS Lett., 577: 170-174 doi:10.1016/j.febslet.2004.10.007 PMid:15527780
Parikh S., Moynihan D.P., Xiao G., and Tonge P.J., 1999, Roles of tyrosine 158 and lysine 165 in the catalytic mechanism of InhA, the enoyl-ACP reductase from Mycobacterium tuberculosis, Biochemistry, 38: 13623-13634 doi:10.1021/bi990529c PMid:10521269
Persson B., Kallberg Y., Oppermann U., and Jörnvall H., 2003, Coenzyme based functional assignments of short-chain dehydrogenases/reductases (SDRs) family, Chem. Biol. Interact., 143-144: 271-278 doi:10.1016/S0009-2797(02)00223-5
Price A.C., Zhang Y.M, Rock C.O., and White S.W., 2001, Structure of β-ketoacyl-[acyl carrier protein] reductase from Escherichia coli: Negative cooperativity and its structural basis, Biochemistry, 40: 12772-12781 doi:10.1021/bi010737g PMid:11669613
Rafferty J.B., Simon J.W., Baldock C., Artymiuk P.J., Baker P.J., Stuitje A.R., Slabas A.R., and Rice D.W., 1995, Common themes in redox chemistry emerge from the X-ray structure of oilseed rape (Brassica napus) enoyl acyl carrier protein reductase, Structure, 3: 927-938 doi:10.1016/S0969-2126(01)00227-1
Schweizer E., and Hofmann J., 2004, Microbial type I fatty acid synthases (FAS): Major players in a network of cellular FAS systems, Microbiol. Mol. Biol. Rev., 68: 501-517 doi:10.1128/MMBR.68.3.501-517.2004 PMid:15353567 PMCid:515254
Sharma S.K., Kapoor M., Ramya T.N.C., Kumar S., Kumar G., Modak R., Sharma S., Surolia N., and Surolia A., 2003, Identiï¬cation, characterization, and inhibition of Plasmodium falciparum {beta}-hydroxyacyl-acyl carrier protein dehydratase (FabZ), J. Biol. Chem., 278: 45661-45671 doi:10.1074/jbc.M304283200 PMid:12930838
Slabas A.R., Chase D., Nishida I., Murata N., Sidebottom C., Safford R., Sheldon P.S., Kekwick R.G.O., Hardie D.G., and Mackintosh R.W., 1992, Molecular cloning of higher-plant 3-oxoacyl-(acyl carrier protein) reductase, Biochem. J., 283: 321-326 PMid:1575676 PMCid:1131036
Smith S., Witkowski A., and Joshi A.K., 2003, Structural and functional organization of the animal fatty acid synthase, Prog. Lipid Res., 42: 289-317 doi:10.1016/S0163-7827(02)00067-X
Tai H.H., Williams M., Iyengar A., Yeates J., and Beardmore T., 2007, Regulation of the β-hydroxyacyl ACP dehydratase gene of Picea mariana by alternative splicing, Plant Cell Rep., 26: 105-113 doi:10.1007/s00299-006-0213-7 PMid:17021849
Yu S., Pan L., Yang Q., Min P., Ren Z., and Zhang H., 2008, Comparison of the Delta (12) fatty acid desaturase gene between high-oleic and normal-oleic peanut genotypes, J. Genet. Genom., 35: 679-685 doi:10.1016/S1673-8527(08)60090-9
. PDF(867KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Xiaoyuan Chi
. Qingli Yang
. Lijuan Pan
. Yanan He
. Mingna Chen
. Yuan Gao
. Shanlin Yu
Related articles
. Peanut ( Arachis hypogaea L.)
. Fatty acid biosynthesis
. β-ketoacyl-ACP reductase (KR)
. β-hydroxyacyl-ACP dehydrase (HD)
. Enoyl-ACP reductase (ENR)
. Expression analysis
Tools
. Email to a friend
. Post a comment


