The Acquisition and Expression Analysis of Polygalacturonase Gene MF6 Related to Fertility in Brassica napus L. 
 Author
Author  Correspondence author
Correspondence author
Genomics and Applied Biology, 2012, Vol. 3, No. 1 doi: 10.5376/gab.2012.03.0001
Received: 25 May, 2012 Accepted: 28 Jun., 2012 Published: 30 Jun., 2012
Huang et al., 2012, The Acquisition and Expression Analysis of Polygalacturonase Gene MF6 Related to Fertility in Brassica napus L., Genomics and Applied Biology, 2012, Vol.3No.1 1-7 (doi: 10.3969/gab.2012.03.0001)
The purpose of this study is to analyze the differentially expressed genes in the buds of male sterile plants which are induced with a male sterilizing chemical "Hua-Sha-Ling WP1" in Brassica napus L., R121. Firstly, we isolated the differentially expressed gene fragments from the male sterile buds through the suppression subtractive hybridization (SSH) and the reverse northern dot blotting. Then, homology comparisons and functional analysis were conducted through online database NCBI. Finally, the differentially expressed gene was further analyzed through reverse transcription quantitative PCR. In the results, we obtained a gene fragment which was 100% homologous to the polygalacturonase gene (MF6) in Arabidopsis from the 6 positive clones. It is reported that the polygalacturonase gene (MF6) is closely related to the development and maturation of pollen. The amount of MF6 gene expression in flower buds with different lengths and different fertilities was analyzed quantitatively. The results showed that the expression of MF6 gene in the male fertile flower buds was higher than that in the male sterile flower buds. And the differences are about 18.53 times and 43.15 times when the bud lengths are 1.00~1.50 mm and 3.5~4.0 mm, respectively. Therefore, "Hua-Sha-Ling WP1" may inhibit the expression of the MF6 gene during the meiosis stages of pollen mother cell and cause the abortion of pollens. At the same time, the MF6 gene expression may also be associated with the elongation of filaments.
Chemical hybridization has become one of the most important ways of utilization of heterosis in crops such as rapeseed, with advantages of thorough male sterilization, freedom in parental selection and combination, freedom from artificial emasculation and so on (Zhang et al., 2011). It can also shorten the breeding period of hybrid cultivars and raise the breeding efficiency (Zhang et al., 2011). Since Naylor (1950) reported that Maleic Hydrazide (MH) could inhibit the development of anther, resulting in male sterility in plants in the 1950s, there have been a large number of scientists who studied the selection, the effects and the mechanism of chemical hybridizing agent (CHA). For example, in the process of male sterility induction by CHA in rapeseed, the release rate of ethylene was increased, the soluble protein content was decreased, the total free amino acid contents were decreased, the free proline content in anthers was decreased, and the enzyme activity was altered (Fan et al., 2008). However, reports on molecular mechanism of the male sterilization by CHA were rare. Studies on the effects of CHA on the expression of relevant genes and the molecular mechanism of plant male sterility will help us with the selection, the synthesis of chemical hybridizing agent in a cheaper cost, a higher efficiency and lower pollution. At the same time, we may also transfer the genes related to male sterilization into superior parents to fasten the breeding progress. In this study, we used an effective CHA, called "Hua-sha-ling WP1", to induce male sterility in a rapeseed line, R121. We used the suppression subtractive hybridization method (SSH) and the reverse northern hybridization technology to display the differentially expressed gene in the flower buds. Homology comparisons and functional analyses were conducted with the identified fragments of genes based on the internet data. The mechanism of male sterility induced by "Hua-sha-ling WP1" was discussed according to the quantitative expressions of the relevant gene.
1 Results and Analysis
1.1 The significant inhibition of "Hua-sha-ling WP1" on the stamen development of Brassica napus L.
The morphology of the sterile flowers was almost the same as that of the fertile flowers, including the size of flowers, the color of petals and the development of pistils, but the filaments were significantly shorter and the anthers were empty compared with the fertile flowers (Figure 1). The rates of sterile flowers were more than 98%.
|
|
1.2 Separation and screening of the differentially expressed fragments in the male sterile flower buds
The products of suppression subtractive hybridization (SSH) were amplified by a second PCR reaction, and connected to pMD19-T vector. After transforming, the screened results indicated that 248 positive clones in the forward subtraction and 359 positive clones in the reverse subtraction were obtained. PCR detections were conducted with a few randomly selected clones. It was shown that the rate of monoclones was higher than 95%. These fragments were clear, and all of them were shorter than 1 000 bp (Figure 2). Two sets of hybridization membrane were prepared for the forward subtractive clones and the reverse subtractive clones. Six positive clones, i.e., 1224, 1252, 1268, 1301, 1319, 1130, were screened out with the reverse northern dot blotting method. These clones were all the expressed fragments of the genes suppressed in the male sterilized flower buds of R121. Figure 3 shows the fragment of No. 1319 screened with the reverse northern dot blotting technology.
|
|
|
|
1.3 Sequencing and homology analyses of the six differentially expressed fragments
The 6 positive clones obtained in the section 1.2 were sequenced and 4 effective cDNA fragments were obtained. The homology and functional analyses were made according to the NCBI database on internet (Table 1). The fragment of No. 1319 was found to be 100% homologous to the sequence of polygalacturonase gene MF6.
|
|
1.4 Quantitative expression analysis of MF6 gene
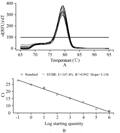
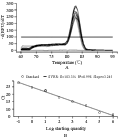
With the results from the section 1.3, the quantitative expression analysis of the MF6 gene in the buds of different fertility and lengths has been done in this experiment. The reference gene UBC21 showed the most stable expression (Table 2), and its amplification specificity and efficiency also achieved the requirements for reference genes by the method of 2-ΔΔCt (Figure 4). The amplification specificity and amplification efficiency of MF6 gene were suitable to use 2-ΔΔCt method for quantitative expression (Figure 5). The expression quantity of MF6 gene in the buds of different fertility and lengths were obtained after the quantitative expression analysis by the 2-ΔΔCt method (Table 3). The expression quantities of MF6 gene in the male fertile buds were apparently higher than in the male sterile buds during the whole development period. The expression quantity of MF6 gene in the male fertile buds was 18.53 times of that in the male sterile buds when the length of buds was 1.00~1.50mm, and 43.15 times when the length of buds was 3.50~4.00mm.
|
|
|
|
|
|
|
|
2 Discussions
Zhang et al (2008) obtained the differentially expressed BcMF6 gene using the cDNA-AFLP technology and the RACE technology in Chinese cabbage (Brassica rapa L.). They constructed an antisense RNA expression vector with the tapetum specific promoter A9, BcA9, and transferred it into the cabbage recipient plants though the Agrobacterium-mediated transformation. They concluded that the BcMF6 gene was pollen specifically expressed PG (polygalacturonase) gene during the maturation process of tapetum and pollens after they detected the morphology after the cytology and the molecular markers were conducted in the transformed Kan-resistant cabbage plants. In this study, we also obtained a cDNA fragment which was 100% homologous to the PG gene MF in the male fertile buds and the male sterile buds induced with a male sterilizing agent "Hua-sha-ling WP1". The results of quantitative expression showed that this gene was highly associated with the fertility performance and bud development in rapeseed (Brassica napus L.). However, when the bud lengths were more than 3.5 mm, and the development of pollens was completed, the expression of MF6 gene was still remained remarkably higher in the male fertile buds than that of the male sterile buds. Therefore, the expression of MF6 gene was not only related to the development of pollens, but might also be involved in the growth and development of other floral organs. According to the results in 1.1, the male sterile flowers had not only empty anthers but also shorter filaments. So MF6 gene may also be associated with the elongation of filaments in Brassica napus L.
Tang et al (2009) observed that the development of pollens was obviously differentiated in the male sterile buds during the period of pollen mother cell stage and the tetrad spore stage in R121 induced by "Hua-sha-ling WP1". In this study, the results showed that the expression of MF6 gene was obviously inhibited in the male sterile buds when the bud length was 1.00~1.50 mm. According to the observation of Gong (2008) on the relationship between bud length and developmental stage of pollens in rapeseed, the development of pollen was at meiosis stage when flower bud was about 1.5 mm in length. This was also an important development stage of tapetum in the rapeseed flower buds. Therefore, the later meiosis stage of pollen mother cells was an important stage for application of "Hua-sha-ling WP1" to induce pollen abortion and produce male sterility in R121. The important period for the application of "Hua-sha-lingWP1" to produce male sterility in R121 could also provide a strong scientific basis and a technical guidance for studies and utilization of chemical hybridizing agents to induce male sterility in Brassica napus L.
3 Materials and Methods
3.1 Materials and reagents
The experimental material R121 is a stable pure B. napus line with normal male fertility, which was provided by the Leshan Academy of Agricultural Sciences, Sichuan Province, China. The male sterilizing agent "Hua-sha-ling WP1" was provided by Fu Yunlong, Mianxian Seed Administration Station, Shanxi Province. The plasmids containing ACTIN, TIP41, UBC21 were supplied by the present laboratory.
Trizol Reagent was purchased from the Invitrogen Company. PCR selectTM and cDNA substraction kit were purchased from the Clontech Company. PMD19-T vector, DNA marker, reverse Taq polymerase, Transcriptase M-MLV, primescript RT reagent kit enzymes (perfect real time), SYBR premix ExTaq â…¡ (perfect real time) were purchased from TaKaRa Company. Plasmid Miniprep kit was purchased from TianGen Company. AxyPrep PCR cleaning kit was purchased from Axygen. Dig High Prime DNA Labeling and Detection Starter Kit â… were purchased from Roche Company.
3.2 Treatments to the materials
On 20th October, 2009, the plant material R121 was planted in 5 rows and 12 hills per row, with 25 cm hill distance on the teaching and research farm of Sichuan Agricultural University. Attention was paid to the control of soil moisture and pest incidence after emergence. Two plants were saved per hill and the extra plants were removed after 2 months. The male sterilizing chemical "Hua-sha-ling WP1" was applied at the initial bolting stage with a concentration of 7 mg/mL. Clean water was sprayed for the first two rows as a control group, and the last two rows were sprayed with the male sterilizing chemical solution, and the central row was used as a spacer row. The spayed amount of solution or water was 9 mL per plant.
3.3 Sampling
Young flower buds were collected with RNA enzyme free tweezers from the initial flowering stage plants. The sample buds were collected from randomly selected male fertile plants and male sterilized plants and kept in liquid nitrogen in freezing tubes. The sample buds were quickly taken to the laboratory and separated into different size groups on ice in an aseptic cabinet in order to meet the requirements of SSH for peer-to-peer system rationality, and to avoid artificial un-uniform abundance and false positive results. The male sterile buds were first divided into five size levels by according to the length: ①0.00~2.00 mm, ②2.00~2.50 mm, ③2.50~3.00 mm, ④3.00~3.50 mm and ⑤3.50~4.00 mm. The fertile buds were also divided into five levels: ⑥0.00~2.00 mm, ⑦ 2.00~2.50 mm, ⑧2.50~3.00 mm, ⑨3.00~3.50 mm, ⑩3.50~4.00 mm. In order to detect the difference in gene expression in a shorter time interval in the quantification analysis of gene expression, the buds of both male sterile plants (A) and male fertile plants (B) were divided into seven size levels, namely, A1 (0.00~1.00 mm), A2 (1.00~1.50 mm), A3 (1.50~2.00 mm), A4 (2.00~2.50 mm), A5 (2.50~3.00 mm), A6 (3.00~3.50 mm), A7 (3.50~4.00 mm); and B1 (0.00~1.00 mm), B2 (1.00~1.50 mm), B3 (1.50~2.00 mm), B4 (2.00~2.50 mm), B5 (2.50~3.00 mm), B6 (3.00~3.50 mm), B7 (3.50~4.00 mm). The divided bud samples were put in freezing tubes, quickly frozen in liquid nitrogen and stored in a -70 ℃ fridge.
3.4 Extraction and detection of the total RNA
The bud samples were extracted using the Trizol Reagent (Cat. No. 15596026) according to the product instructions. The quality of the total RNA was detected by agarose gel electrophoresis, and the purity and the concentration of RNA were examined with a nucleic acid and protein detecting instrument. When the 28S and 18S bands were clearly seen and the ratio of OD260/OD280 was around 2.0, further experiments were continued.
3.5 Suppression subtractive hybridization
Total RNA of the five levels (①~⑤) of buds from the male sterile plants was mixed into sample 1 (R121 male sterile buds), and the total RNA of the five levels (⑥~⑩) of buds from the male fertile plants was mixed into sample 2 (R121 male fertile). A forward substractive hybridization was conducted using the sample 1 as tester and the sample 2 as driver, and a reverse substractive hybridization was conducted using the sample 2 as tester and the sample 1 as driver, following the product instructions of Subtraction Kit PCR-SelectTM (Cat. No. 637401).
3.6 Construction of subtracted cDNA library and PCR detection
After a second time PCR amplification the final products of the forward and the reverse subtractive hybridization were purified using the PCR Cleaning Kit (Cat. No. AP-PCR). Cloning and transformation were made following the instruction of pMD19-T vector (Cat. No. D102A) of the Takara Company. A forward subtractive cDNA library and a reverse subtractive cDNA library were constructed, and the products were kept in tubes at -70℃ after loading 200 μL glycerol in the tubes.
Thirty tubes of bacteria suspension were randomly taken from the forward subtractive library and the reverse subtractive library. PCR amplification was performed using 1 μL of the bacterium template DNA, together with the nested primer l and the nested primer 2R (provided in the reagent box) as primers, in a 20 μL reaction system. The reaction conditions were the following: predenaturing at 94℃for 20 s; followed by 27 cycles of denaturing at 94℃for 30 s, annealing at 68℃for 30 s, and extension at 72℃for 1 min; and a final extension at 72℃for 10 min; finally stored at 12℃ forever.
The rates of monoclones were calculated and the fragment sizes of the clones were identified using 1% agarose gel electrophoresis.
3.7 Screening of the positive clones and sequencing
The sample 1 and sample 2 RNA’s obtained in the section 1.2.4 were reversely transcribed into the first strand of cDNA, according to the specification of the reverse transcriptase enzyme, M-MLV (Cat. No. D2639, Takara Co.). The products were labeled R121A and R121B, respectively, with digoxigenin according to the Roche DIG High Prime DNA Labeling and Detection Starter Kit â… (Cat. No. 11745832910). Two same sets of hybridization membrane were prepared for the subtractive cDNA libraries. One set was used to hybridize with the probe R121A, and another was used to the probe R121B after they were baked and pre-hybridized. The two sets of membrane were hybridized over night at 42℃, and were washed and immunologically colorized for comparison. The positive clones on the membrane of the forward subtractive library were recognized when color was observed with the probe R121A, but not with probe R121B or a deeper color was observed with probe R121A than with probe R121B. The positive clones on the membrane of reverse subtractive library were recognized when color was observed with probe R121B, but not with probe R121A, or the color with probe R121B was deeper than with probe R121A. The positive clones were sequenced by the Invitrogen Co, Shanghai.
3.8 Homology and function analyses of the sequences
The data of sequences from the section 3.7 were put into the software DNAMAN, and the vector and linker sequences were removed. The cDNA sequences that we obtained were submitted to the NCBI database (http://blast.ncbi.nlm.nih.gov/ Blast.cgi ) for an online homology comparison. The functions of the genes were predicted based on the reference annotation they well matched and on literature.
The software Primer 5 was used to design the primers for the quantitative expression analyses based on the above results.
3.9 Fluorescent quantitative expression analysis of the functional gene
3.9.1 Synthesis of the first strand of cDNA
The total RNA from the male sterile buds (A1, A2, A3, A4, A5, A6, A7) and the male fertile buds (B1, B2, B3, B4, B5, B6, B7) in the section 3.4 were reversely transcribed into the first strands of cDNA according to the specification of the PrimeScript RT reagent Kit. (Perfect Real Time, Cat. No. DRR037S).
3.9.2 Selection of reference gene and detection of amplification efficiency
The reversely transcribed cDNA from different lengths of buds was used as template. Primers were from the generally used internal reference gene actin (F: TCCTCACGCTATCGCTATCCT CCG; R: GATGTTTCCATACAGATCCTTCC) and two reported internal reference gene, tip41 (F: AGAGTCATGCCAAGTTCATGGTT; R: CCTCATAAGCACACCATCAACTCTAA), and ubc21 (F: CCTCTGCAGCCTCCTCAAGT; R: CATATCTCCCCTGTCTTGAAATCG) in literature (Chen et al., 2010), respectively. The least variable and stably expressed internal reference gene was screened out following the operation manual of the SYBR Premix Ex Taq â…¡ Perfect Real Time (Cat. No. DRR081S) by TaKaRa.
In order to determine the amplification efficiency and specificity of the reference gene, the plasmid containing the reference gene was diluted by multiples of 10 into a series of gradient dilution to the concentration of 10-8. Each gradient plasmid dilution was used as template to conduct Real-Time quantitative PCR, sequentially, in a 15 μL volume system on the PCR instrument BIO-RAD CFX96. Dissolution curve and standard curve of the reference gene were obtained using the CFX manager software provided with the BIO-RAD CFX96 instrument.
3.9.3 Extraction of plasmid DNA and test of the primer amplification efficiency
The bacterium clone with the target gene fragment was further multiplied. Plasmid of this clone was extracted according to the plasmid Miniprep Kit (Cat. No. DP10302). Then a set of gradient dilution of the plasmid were prepared in multiples of 10 to the concentration of 10-8. The gradient plasmid dilutions were used as template sequentially. The primers for the target gene were designed with the software Primer 5. Real-time fluorescent quantitative PCR was performed in a 15 μL volume system on the instrument BIO-RAD CFX96 PCR. The primer amplification efficiency and specificity were analyzed and the dissolution curve and standard curve of the target gene were obtained using the CFX manager software provided with the BIO-RAD CFX96 instrument.
3.9.4 The expression of MF6 gene in the buds with different fertility and length
The first strand of cDNA transcribed from the total RNA of the 7 grades of male fertile and male sterile buds was used as templates. The primers were those previously screened for the internal reference genes and the target gene. The reaction system and the PCR procedure were the same as used in the establishment of the standard curve. The CFX manager software with the PCR instrument BIO-RAD CFX96 was used to analyze the expression of the target gene MF6 when the reaction was completed.
Authors’ Contribution
RXH participated the experimental design, conducted the experiment operation, data analysis and paper writing. ZKT and SXG participated in the experimental design, provided the plant materials, and gave experiment instruction. YZN, the responsible author, provided the project idea, directed the experimental design, operation and data analyses. All authors have read and approved the final manuscript.
Acknowledgements
This study was funded by the National “Twelfth Five-year Plan” Projects for the Rural Development (2011AA10A104) and the Sichuan Province Rapeseed Breeding Key Projects (2011-2015). All the authors appreciated two anonymous reviewers for their useful critical comments and revising advice to this paper.
Reference
Chen X., Truksa M., Shah S., and Weselake R.J., 2010, A survey of quantitative real-time polymerase chain reaction internal reference genes for expression studies in Brassica napus L., Analytical Biochemistry, 405(1): 138-140
http://dx.doi.org/10.1016/j.ab.2010.05.032PMid:20522329
Fan B.L., Yue X.L., Zheng Q., and Meng X.K., 2008, Study on the SOD, CAT and POD activity in leaves and buds of rape induced by WP and YB, Hubei Nongye Kexue (Hubei Agricultural Sciences), 47(4): 406-408
Gong L., 2008, Studies on relationship between length of flower buds and pollen development stages in rape, Anhui Nongye Tongbao (Auhui Agricultural Science Bulletin), 14(17): 61-63
Naylor A.W., 1950, Observations on the effects of maleic hydrazide on the f1owering of tobacco, maize and cocklebur, Proc. Natl. Acad. Sci., USA, 36(4): 230-232
http://dx.doi.org/10.1073/pnas.36.4.230
Tang Z.K., 2009, Study on the male abortion induced by a chemical hybridizing agent “Huashaling” in rapeseed (Brassica napus L.), Thesis for M.S., Sichuan Agricultural University, Supervisor: Niu Y. Z., pp.30-33
Zhang Q., Huang L., Liu T.T., Yu X.L., and Cao J.S., 2008, Functional analysis of a pollen-expressed polygalacturonase gene BcMF6 in Chinese cabbage (Brassica campestris L. ssp. Chinensis Makino), Plant Cell Reports, 27(7): 1207-1215
http://dx.doi.org/10.1007/s00299-008-0541-xPMid:18415101
Zhang Z.Q., Wang G.H., Guan C.Y., and Chen S.Y., 2011, Research advances in chemical emasculation of rape, Hunan Nongye Kexue (Hunan Agricultural Sciences), 5: 19-22
. PDF(404KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Rongxian Huang
. Zhikang Tang
. Shixing Guo
. Yingze Niu
Related articles
. Brassica napus L.
. Chemical male sterilization
. Differentially expressed genes
. MF6
. Pollen abortion
Tools
. Email to a friend
. Post a comment