Research Article
Function Analysis of cDNA Clone Genes from Puccinellia tenuiflora under NaHCO3 Stress 
2. Institute of Life Science, Jiyang College of Zhejiang A&F University, Zhuji, 311800, China
 Author
Author  Correspondence author
Correspondence author
Molecular Soil Biology, 2016, Vol. 7, No. 9 doi: 10.5376/msb.2016.07.0009
Received: 25 Mar., 2016 Accepted: 12 Apr., 2016 Published: 12 Apr., 2016
Liu P.P., Sun Y.J., Ye X.X., and Fang J.X.J., 2016, Function Analysis of cDNA Clone Genes from Puccinellia tenuiflora under NaHCO3 Stress, Molecular Soil Biology, 7(9): 1-16 (doi: 10.5376/msb.2016.07.0009)
Puccinellia tenuiflora is widely found in saline-alkali soil of the Songnen plain in northeastern China, which indicates it has a high tolerance to saline-alkali stress. In this study, we selected 3,072 clones from cDNA library of P. tenuiflora with NaHCO3 treatment. In the end, 2991 clones were chosen for subsequent analysis. We use blastx to explore the homology relationship between P. tenuiflora and other plants. Among 2991 clones, approximately 89% were annotated in more than one public protein database and nearly 25% of the clones showed significant similarities with Hordeum vulgare. The clone sequences are classified in carbohydrate metabolism, photosystem, response to stimulus, ribosome, defense mechanism, metal/ion binding and hormone-mediate signal, etc. And from the 2991 clones, we identified 184 clones which may be involved in salt stress by search pubmed database with corresponding keywords. This study offers new molecular functions and mechanisms of P. tenuiflora and may also provide valuable reference for exploring potential molecular mechanisms of saline-alkali tolerance in P. tenuiflora.
Introduction
Saline soil is defined as soil that having electrical conductivity of 4 dS m−1 (Chinnusamy et al., 2005). According to incomplete UNESCO and FAO statistics, the world of saline-alkali land area is around 954.38 million hectares (Flowers, 2004; Peng et al., 2004). And in China there is about 99.13 million hectares (G.X et al., 2003; Mackerness et al., 2001). The causes of soil alkalinity are natural or they can be man-made. In China, the formation of saline-alkali soils is associated with the cumulative of carbonates in the soil. Because of the low infiltration capacity, rain water stagnates, and in dry periods, cultivation is hardly possible without copious irrigated water and good drainage. Salinity stress will hamper plant growth in terms of osmotic stress, ion toxicity, ROS-induced oxidative stress, nutrient deficiency and hormonal imbalance (Ashraf M, 2004; Ashraf and Akram, 2009). So saline-alkali soils are difficult to take into agricultural production (Ahmad, 2010). To improve the agricultural potential of these saline-alkali soils, at present there are only two ways to solve this. One is to improve natural conditions by men, such as fresh watering irrigation; the other method is to cultivate glasswort or saltwort or barilla plants such as screening the crops that can adapt to heavy salt environment (Chunxue et al., 2008). At present, it is more practical to study and explore more salt tolerance plants than artificial works. But saline-alkali soils are difficult to grow, and only a few kinds of plants can survival in saline-alkali soil. Breeding transgenic plants that are tolerant of saline-alkali soil and the study of salt tolerance plants on the molecular level have important significance. It may be a reference to improve plant salt tolerance, and make the effective utilization of saline soil.
Previous Studies have shown that some general molecular mechanisms of salt tolerance are involved in pathways related to Ion transport (Peng et al., 2004; Pons et al., 2011; Wang et al., 2013), SOS pathway (Mahajan and Tuteja, 2005) , ABA (abscisic acid) and MKK2 pathway , glycine betaine (D Rhodes and Hanson, 2003), mitogen-activated protein kinases (Vashisht and Tuteja, 2006b; Zhang et al., 2006), DEAD-box helicases (Vashisht and Tuteja, 2006a), proline (Thiery et al., 2004) and reactive oxygen species (Miller et al., 2008). These pathways play major roles in salt tolerance. However, these researches were all studied under the stress of NaCl; we still know little about stress-response proteins in plants under saline-alkali stress such as HCO3- and CO32-.
Different plants to saline-alkali stress vary greatly in their different growth stage. Most Crops are glycophytes which can’t grow under high-saline conditions. In the other hand, the halophytic plants have evolved some mechanisms to hamper salt stress (Ardie et al., 2010; Flowers and Colmer, 2008). Studying about halophytic plants on molecular level will help us to understand the vital mechanisms of salt tolerance and explore some interesting candidate genes. Here, we use Puccinellia tenuiflora as study object to study the mechanisms and pathways involved in salt tolerance and improve the salt tolerance of cereals. Puccinellia tenuiflora are halophytic species belonging to the genus Gramineae, known as alkali grass or salt grass (Liu et al., 1848; Wang et al., 2007). Puccinellia tenuiflora is widely found in saline-alkali soil of the Songnen plain in northeastern China and used as forage for soil improvement (Wang et al., 2009). Previous studies have shown that the mechanism of P. tenuiflora is due to salt secretion (Yan et al., 1994). Another suggestion of P. tenuiflora is that the endodermis hider the Na+ absorption while absorption of K+ is maintained (Peng et al., 2004). In 2007, researchers have identified EST in alkali grass by constructing cDNA library and EST analysis (Wang et al., 2007). However, the mechanism of salt tolerance of P. tenuiflora is still not clear.
In this study, we want to explore the global gene expression patterns of P. tenuiflora with NaHCO3 treatment. We constructed a cDNA library of P. tenuiflora and EST analysis to analysis the response of P. tenuiflora to alkali salt at the level of genomic. Use homology analysis and function annotation of clone sequences with known genes by blastx, 89% were annotated in more than one publicly available protein database. Based on the matches in Nr, COG, GO and KEGG, most clones were classified in basic biological processes, Energy production and conversion, response to stimulus, ion transporter, membrane transport and ribosome, plant hormone signal transduction and other cellular components and function. And from the results of annotation, we identified 184 clones that may have a role in the salt tolerance such as Na+/H+ exchanger (S04106_27_C03 with p-value = 6.03E-123), ammonium transporter (S04106_26_F09 with p-value 3.21E-138) with an important role in slat tolerance in A. thaliana. So the study may provide a useful reference in exploring candidate saline-alkali tolerance genes and Breeding transgenic plants that are tolerant of saline-alkali soil.
Materials and Methods
Plant material, culture condition and stress condition
The P. tenuiflora were collected from the Anda experimental base of Alkali Soil Natural Environmental Science C enter of Northeastern Forestry University. Plants were germinated in tap water with distilled water for 2-3 weeks under controlled conditions of 60% relative humidity, 16 light (6,000 Lux) daily and an average temperature of 24℃. Then the plants were treated with 150 mmol NaHCO3 instead of distilled water for salt-stress. After 5 days’ treatment, young roots were harvested and immediately frozen in -80℃ for future analysis.
RNA extraction and cDNA library construction
Total RNA samples were extracted from NaHCO3 treated P. tenuiflora roots using Trizol reagent (Invitrogen Corp., Carlsbad, CA) according to the manufacturer’s instructions, purified using the QIAGEN RNeasy Kit (Qiagen Inc., Valencia, CA). The RNA samples were used to construct cDNA library of P. tenuiflora with SMART cDNA Library Construction Kit according to the manufacturer’s instructions.
Sequences processing and quality control
A total number of 3,072 clones were single-pass sequenced from their 5’-end and filter these sequences with following criteria: (1) the length of clone sequences more than 100bp, (2) the Enzyme loci of SwaI Enzyme (ATTT^AAAT), (3) the quality of sequences.
Function annotation and analysis of clone sequences
BLAST alignment was performed with a typical cut-off E-value of 10-5 between sequences, using publically available protein data from several databases (Johnson et al., 2008): NCBI non-redundant protein (NR), Clusters of Orthologous Groups (COGs) and KOG database (Galperin et al., 2015). The best blastx outputs were used to infer function of the sequences. We use KAAS (Moriya et al., 2007) (http://www.genome.jp/tools/kaas/) to analysis functional annotation of genes by BLAST comparisons against the manually curated KEGG GENES database (Kanehisa et al., 2015). Blast2go (Conesa and Gotz, 2008) program with NR annotation was used to get GO annotations at category of biological processes, molecular functions, and cellular components ontologies (balst2go ref). Then we use WEGO to display GO annotation classification of 2,846 sequences, to explore the distribution of gene function of sequences at GO level (WEGO ref). With the results of blastx against NR database, we use the hit accession number of blast best hit of each sequence to annotate gene function with DAVID (Huang et al., 2009a; Huang et al., 2009b).
Results and Discussion
Homology analysis and function annotation of clone sequences
After heading and tailing the vectors, poor quality sequences and sequences shorter than 100 bases were removed, and the final number of clone sequences was 2,991. We make annotation for the 2,991 clone sequences by searching database Non-redundant protein sequences (nr)(ftp://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/nr.gz)using Blastx with an E-value cut-off of 1.0E−5. These sequences alignments results were used to retrieve proteins with the highest sequence similarity to the P. tenuiflora clone sequences to infer their functional annotations. Among 2,991 sequences that we selected from P. tenuiflora, 2620 of them about 87.60% had significantly blast hits in NCBI non-redundant protein database (NR database) (Fig.1.A). The absence of blast hits in public databases may indicate that novel genes are specifically expressed in P. tenuiflora, or it may be due to other biological or technical biases.
|
Figure 1 Statistics of Blast2GO project data and distribution of Blastx search for clone sequences against NR database. Fig1.A represents the statistics of Blast2GO project data. 371 of them had no blast hits, whereas 87.5% of clone genes had blast hits. Among these clone sequences, 1590 clone sequences had GO (GO-slim) annotation. (B) The distribution of species for the top BLAST hits for each clone sequence (with an E-value ≤1.0E-5). (C) The distribution of similarity is shown as the percentage of the total homologous sequences (with an E-value ≤1.0E-5) (D) E-value distribution of BLAST hits for clones sequences with an E-value cut-off of 1.0E-5. During the homology searching, we used all proteins in NR database, and the best hits of clone sequences were used for further analysis. |
From the results of Blastx with NR database, we found that the species distribution of the top hits in the NR database showed that most of the mapped sequences had strong homology with plants, whereas 25% of the sequences were annotated to Hordeum vulgare, followed by Brachypodium distachyon (22%), Aegilops tauschii (10%), Triticum aestium (7%) and Tritium urartu (7%) (Fig.1.B). among of the top ten species of blastx hits, eight are belonging to angiospermae. The distribution of identity values showed that 48 % of the query sequences had similarity score with Blastx hits more than 90%, whereas 46% of the blast hits had a similarity ranging from 70% to 90% (Fig.1.C). The E-value distribution of BLAST hits for matched sequences showed that 79% of the mapped sequences had strong similarity with the E-value <1.0E-50. And 21% of the homolog sequences ranged from 1.0E-50 to 1.0E-5 (Fig.1.D). The Blastx results indicated that 2,620 (87.6% of all clone genes) had good comparability to known gene sequences, and the statistics of data indicated the clone sequences were annotated properly. We selected the best Blastx hit of each clone sequence from Blastx results for further analysis.
|
Figure 2 Histogram representations of GO classification and function annotation with DAVID. Fig.2A represented the results of GO annotation were classified in three main categories: cellular component, molecular function and biological process. And B displayed the gene function annotation chat of DAVID analysis results. |
Classification of Gene ontology (GO) and DAVID gene annotation
With the results of Blastx against NR database, Blast2GO was used for GO annotation based on biological processes, molecular functions, and cellular components ontologies. From the BLAST2GO annotation results, we found that 53.2% of clone sequences were annotated to GO terms (Fig1.A). These 1,590 out of 2,847 clone sequences were categorized into 33 functional groups at GO level 2 that belong to the biological process, molecular function and cellular component clusters. Using WEGO (http://silkworm.swu.edu.cn/cgi-bin/wego/index.pl) to display GO annotation results of GO level 2 (Fig.2A). Among 2,991 clone sequences, 1,346 clone sequences were in the category of biological processes, 946 clone sequences were annotated in the category of cell component and 1,328 of these were classified in the category of molecular function. In the biological processes category, 1,346 clone sequences mapped to 18 terms including “metabolic process”, “cellular process”, “single-organism process”, “localization”, “response to stimulus”, “cellular component organization or biogenesis” and so on. And the dominant subcategories were “cell” and “cell part” in cellular components, and among molecular functions, the major subcategories is “binding” and “catalytic”.
To better understand and explore the functions of these clone sequences, we did function annotation analysis with DAVID. Here, using accession number of best hit of each clone sequence to do DAVID analysis. Among the 2991 clone sequences, 14 are mapped to UniProtKB, 66 are mapped to GenePept accession, and 49 are mapped to Refseq-protein. Then using the 129 mapped sequences do gene function analysis with DAVID tools. As the results of Blastx with NR, the top similarity species is Hordeum vulgare (Fig.1.B); we use Hordeum vulgare as the background gene set to do annotation. From With the high classification stringency, we got 4 functional annotation clusters including photosystem, chloroplast and transit peptide, ion binding and transmembrane (Table 1). As known, general molecular mechanisms of salt tolerance are involved in pathways related to Ion transport. In the result of DAVID, we found the genes are classified in the terms of transit peptide, ion transporter and transmembrane which have a close relationship with ion transporter and membrane transport (Pons et al., 2011). So the clone sequences that were involved in these terms may play a role in salt tolerance.
As most of genes in Hordeum vulgare are predicted genes, and salt stress studies about Oryza sativa are more than Hordeum vulgare, we did another DAVID analysis with Oryza sativa as background gene set. From the results of DAVID analysis, we found that the clone sequences commonly are classified into “protein transport”, “hormone-mediated signaling”, “cellular response to hormone stimulus”, “response to endogenous stimulus” (Fig.2.B). Previous studies have shown that plant hormones including abscisic acid (ABA) (Hadiarto and Tran, 2011; Raghavendra et al., 2010; Ryu and Cho, 2015; Xiong et al., 2002; Zhu 2002), auxin, cytokinins (CK) (Hwang and Sheen, 2001; Javid et al., 2011; Mok and Mok, 2001; Ryu and Cho, 2015), brassinosteroids (BRs) (Kang et al., 2005; Kazan and Manners, 2012; Moons et al., 1997; Ryu and Cho, 2015), jasmonate, gibberellin (GA) (Daviere and Achard, 2013; Ryu and Cho, 2015; Sun and Gubler, 2004) and ethylene show important role in salt tolerance in plants. So the clone sequences in these terms may play a major role in salt stress of P. tenuiflora. S04106_06_H05.seq was annotated as Auxin response factor 9 which has an important role in regulating plant growth and development and several evidences involvement of auxin in response to salinity stress in plants.
COG function classification and KEGG pathway annotation
The cluster of orthologous groups (COG) is used to classify orthologous gene products. The database contains the putative protein sequences encoded by all genes from bacteria, archaea and eukaryotes with completed genome sequences and explores the evolutionary relationships between these groups through sequence analysis. Newly unknown sequences can be aligned to the COG database and KOG database to predict and infer their possible functions by blastx with a cut-off e-value = 1E-5. A total of 1,125 clone sequences were annotated into 24 molecular families. The function class for “Carbohydrate transport and metabolism” (201, 17.87%) represents the largest group among the 24 COG categories, followed by “translation, ribosomal structure and biogenesis” (174, 15.46%), “posttranslational modification, protein turnover, chaperones” (143, 12.71%), “General function prediction only” (109, 9.69%) and “Energy production and conversion” (100, 8.89%) (Figure3).
|
Figure 3 Distribution of COG annotations for clone sequences. The horizontal axis represents COG function family, and the vertical axis showed how many clone genes were classified into the corresponding COG function class. |
|
Table 1 DAVID function annotation clusters of mapped sequences Annotation Cluster 1 Enrichment Score: 1.5568309284413362 |
To identify the biological pathways of the clone sequences of P. tenuiflora, we analyze the 2,991 with KEGG Automatic Annotation Server for orthologous assignment and pathway mapping. In total, 1847 clone sequences were assigned to 295 KEGG pathways. The pathways with most representation by the unique sequences were metabolic pathways (200 members) and Biosynthesis of secondary metabolites (98 members) and plant hormone signal transduction. Plant hormones have an important role in alleviation of salt stress in plant (Ryu and Cho, 2015). Here, plant hormone signal transduction pathway was displayed as Figure 4. By KAAS results, the clones of S04106_28_A09, S04106_15_G05, S04106_06_H05 were assigned as AUX1 (LAX auxin influx carrier), auxin-responsive protein IAA, and ARF (auxin response factor). In previous studies, researchers have suggested that function of some Aux/IAA genes were related to salt stress (Wang et al., 2010). While S04106_14_B01 was annotated as EIN3 (ethylene-insensitive protein 3), which have complex regulatory roles during abiotic stress adaptation (Kazan, 2015). In brief, all these annotations provide a valuable resource for investigating specific functions, processes, and pathways associated with molecular mechanisms of saline-alkali stress in P. tenuiflora In brief, all these annotations provide a valuable resource for investigating specific functions, processes, and pathways associated with molecular mechanisms of saline-alkali stress in P. tenuiflora.
|
Figure 4 the KAAS of results of plant hormone signal transduction. The green genes/proteins are the query clones assigns KO identifiers. Then in the assigned pathway mapping the KO identifies are colored green. |
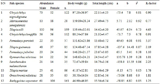
To identify potential saline-alkali stress associated genes in the 2,991 clones, by searching NCBI PubMed database with the keywords “salt stress”, “plant” and corresponding annotation of each clones, 185 clones were identified. For the 184 clones, we collected the id, database, the accession number, description, species, p-value and reference article PMID (supplementary material table S1). In table 2, we displayed the first 10 records of the clones. Most of the clones were annotated to H+ ATPase, metal transporters, ammonium transporter, CBS-domain, glutathione S-transferase and aminotransferase. It was suggested that the clones S04106_03_F10 (auxin-repressed protein), S04106_15_G05 (auxin-responsive protein IAA24-like), S04106_16_A03 (auxin-repressed 12.5 kDa protein-like), S04106_28_A09 (LAX auxin influx carrier) which take part in plant hormone signal transduction pathway (Figure 4) may play an important role in abiotic stress adaptation (Wang et al., 2010). In the 185 clones, some were annotated as transcription factor such as S04106_02_B06 (MADS-box transcription factor), S04106_18_A08 (myb family transcription factor), S04106_18_F04 (Heat stress transcription factor) and S04106_31_F07 (probable WRKY transcription factor). Previous studies showed that MADS-box genes may be involved in abiotic stress was mentioned in the research of Wei et al (Wei et al., 2014). These potential clones may have an important role in saline-alkali tolerance.
|
Table 2 clones may be involved in saline-alkali tolerance |
Conclusion
In this study, we randomly selected 3072 clones from cDNA library of P. tenuiflora with NaHCO3 treatment. We finally selected 2991 clones for subsequent analysis. Use homology analysis and function annotation of clone sequences with known genes by blastx, 89% were annotated in more than one publicly available protein database. Based on the matches in Nr, COG, GO and KEGG, most clones were classified in basic biological processes, Energy production and conversion, response to stimulus, ion transporter and membrane transport, plant hormone signal transduction and other cellular components and function. The function analysis of these clones of P. tenuiflora with NaHCO3 treatment offered valuable information about the saline-alkali tolerance in P. tenuiflora. With homology search, some clones in P. tenuiflora such as S04106_28_A09, S04106_15_G05, and S04106_06_H05 et al. may associate with saline-alkali responsive were identified. We also identified 185 clones by searching NCBI PubMed database with keywords “salt stress”, “plant” and corresponding annotation of each clone. These candidate clones may have important role in saline-alkali tolerance, and facilitate breeding transgenic plants that are tolerant of saline-alkali soil.
References
Farooq Ahmad. (2010). Leptochloa Fusca Cultivation for Utilization of Salt – affected Soil and Water Resources in Cholistan Desert. Sociedade & Natureza, Uberlândia, 22(1), 141-149.
http://dx.doi.org/10.1590/S1982-45132010000100010
S. W. Ardie, S. Liu, & T. Takano. (2010). Expression of the AKT1-type K(+) channel gene from Puccinellia tenuiflora, PutAKT1, enhances salt tolerance in Arabidopsis. Plant Cell Rep, 29(8), 865-874. doi: 10.1007/s00299-010-0872-2
http://dx.doi.org/10.1007/s00299-010-0872-2
Ashraf M. (2004). Some important physiological selection criteria for salt tolerance in plants. Flora of China, 199(5), 361-379.
http://dx.doi.org/10.1078/0367-2530-00165
M Ashraf , & NA Akram. (2009). Improving salinity tolerance of plants through conventional breeding and genetic engineering: an analytical comparison. Biotechnol Adv, 27(6), 744-752.
http://dx.doi.org/10.1016/j.biotechadv.2009.05.026
Viswanathan Chinnusamy, Andre´ Jagendorf, & Jian-Kang Zhu. (2005). Understanding and Improving Salt Tolerance in Plants. CROP SCIENCE, 45(2), 437-448.
http://dx.doi.org/10.2135/cropsci2005.0437
Yang Chunxue, Luo Qiuxiang, Zhuo Lihuan, & Liu Shenkui. (2008). Preliinary Identification of Root Specific-Expressed Protein in Puccinellia tenuiflora under NaHCO3 Stress. Molecular Plant Breeding, 6(4), 669-674.
A Conesa, & S. Gotz. (2008). Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics, 2008, 12. doi: 10.1155/2008/619832
http://dx.doi.org/10.1155/2008/619832
D Rhodes, & AD Hanson. (2003). Quaternary ammonium and tertiary sulfonium compounds in higher-plants. Plant Mol. Biol., 44, 357-384.
JM Daviere , & P Achard. (2013). Gibberellin signaling in plants. Development, 140(6), 1147-1151. doi: 10.1242/dev.087650
http://dx.doi.org/10.1242/dev.087650
T. J. Flowers. (2004). Improving crop salt tolerance. Journal of Experimental Botany, 55(396), 307-319. doi: 10.1093/jxb/erh003
http://dx.doi.org/10.1093/jxb/erh003
T. J. Flowers , & T. D. Colmer. (2008). Salinity tolerance in halophytes. New Phytologist, 179(4), 945-963. doi: 10.1111/j.1469-8137.2008.02531.x
http://dx.doi.org/10.1111/j.1469-8137.2008.02531.x
Dai G.X, Peng K.Q, & Pi C.H. (2003). The Effects of Calcium on Salt-tolerance in Plant. CHINESE AGRICULTURAL SCIENCE BULLETIN, 19(3), 97-101.
MY Galperin, KS Makarova, YI Wolf, & EV Koonin. (2015). Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res, 43(Database issue), D261-269. doi: 10.1093/nar/gku1223
http://dx.doi.org/10.1093/nar/gku1223
T. Hadiarto , & L. S. Tran. (2011). Progress studies of drought-responsive genes in rice. Plant Cell Rep, 30(3), 297-310. doi: 10.1007/s00299-010-0956-z
http://dx.doi.org/10.1007/s00299-010-0956-z
DW Huang, BT Sherman, & RA Lempicki. (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res, 37(1), 1-13. doi: 10.1093/nar/gkn923
http://dx.doi.org/10.1093/nar/gkn923
DW Huang, BT Sherman, & RA Lempicki. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols, 4(1), 44-57. doi: 10.1038/nprot.2008.211
http://dx.doi.org/10.1038/nprot.2008.211
I. Hwang , & J. Sheen. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature, 413(6854), 383-389. doi: 10.1038/35096500
http://dx.doi.org/10.1038/35096500
M. G. Javid, A. Sorooshzadeh, F. Moradi, S. A. M. M. Sanavy, et al. (2011). The role of phytohormones in alleviating salt stress in crop plants. Australian Journal of Crop Science, 5(6), 726-734.
M. Johnson, I. Zaretskaya, Y. Raytselis, Y. Merezhuk, et al. (2008). NCBI BLAST: a better web interface. Nucleic Acids Res, 36(Web Server issue), W5-9. doi: 10.1093/nar/gkn201
http://dx.doi.org/10.1093/nar/gkn201
M Kanehisa, Y Sato, M Kawashima, M Furumichi, et al. (2015). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res, 35(2), W182-185. doi: 10.1093/nar/gkv1070
http://dx.doi.org/10.1093/nar/gkv1070
DJ Kang, YJ Seo, JD Lee, R. Ishii, et al. (2005). Jasmonic acid differentially affects growth, ion uptake and abscisic acid concentration in salt-tolerant and salt-sensitive rice cultivars. Journal of Agronomy and Crop Science, 191(4), 273-282. doi: DOI 10.1111/j.1439-037X.2005.00153.x
http://dx.doi.org/10.1111/j.1439-037X.2005.00153.x
K Kazan , & JM Manners. (2012). JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci, 17(1), 22-31. doi: 10.1016/j.tplants.2011.10.006
http://dx.doi.org/10.1016/j.tplants.2011.10.006
K. Kazan. (2015). Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci, 20(4), 219-229. doi: 10.1016/j.tplants.2015.02.001
http://dx.doi.org/10.1016/j.tplants.2015.02.001
Liang Liu, Guanghua Zhu, & Nikolai N. Tzvelev. (1848). Puccinellia Parlatore. Flora of China, 22, 1, 225, 245, 287, 315.
AH Mackerness, C. F. John, B. Jordan, & B. Thomas. (2001). Early signaling components in ultraviolet-B responses: distinct roles for different reactive oxygen species and nitric oxide. FEBS Lett, 489(2-3), 237-242.
http://dx.doi.org/10.1016/S0014-5793(01)02103-2
S. Mahajan , & N. Tuteja. (2005). Cold, salinity and drought stresses: an overview. Arch Biochem Biophys, 444(2), 139-158. doi: 10.1016/j.abb.2005.10.018
http://dx.doi.org/10.1016/j.abb.2005.10.018
G. Miller, V. Shulaev, & R. Mittler. (2008). Reactive oxygen signaling and abiotic stress. Physiol Plant, 133(3), 481-489. doi: 10.1111/j.1399-3054.2008.01090.x
http://dx.doi.org/10.1111/j.1399-3054.2008.01090.x
DW Mok , & MC Mok. (2001). Cytokinin Metabolism and Action. Annu Rev Plant Physiol Plant Mol Biol, 52(4), 89-118. doi: 10.1146/annurev.arplant.52.1.89
http://dx.doi.org/10.1146/annurev.arplant.52.1.89
A. Moons, E. Prinsen, G. Bauw, & M. Van Montagu. (1997). Antagonistic effects of abscisic acid and jasmonates on salt stress-inducible transcripts in rice roots. Plant Cell, 9(12), 2243-2259. doi: 10.1105/tpc.9.12.2243
http://dx.doi.org/10.1105/tpc.9.12.2243
Y Moriya, M Itoh, S Okuda, AC Yoshizawa, et al. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res, 35(Web Server issue), W182-185. doi: 10.1093/nar/gkm321
http://dx.doi.org/10.1093/nar/gkm321
YH Peng, YF Zhu, YQ Mao, SM Wang, et al. (2004). Alkali grass resists salt stress through high [K+] and an endodermis barrier to Na+. Journal of Experimental Botany, 55(398), 939-949. doi: 10.1093/jxb/erh071
http://dx.doi.org/10.1093/jxb/erh071
R Pons, MJ Cornejo, & A Sanz. (2011). Differential salinity-induced variations in the activity of H(+)-pumps and Na(+)/H(+) antiporters that are involved in cytoplasm ion homeostasis as a function of genotype and tolerance level in rice cell lines. Plant Physiol Biochem, 49(12), 1399-1409. doi: 10.1016/j.plaphy.2011.09.011
http://dx.doi.org/10.1016/j.plaphy.2011.09.011
AS Raghavendra, VK Gonugunta, A Christmann, & E. Grill. (2010). ABA perception and signalling. Trends Plant Sci, 15(7), 395-401. doi: 10.1016/j.tplants.2010.04.006
http://dx.doi.org/10.1016/j.tplants.2010.04.006
Hojin Ryu , & YongGu Cho. (2015). Plant hormones in salt stress tolerance. Journal of Plant Biology, 58(3), 147-155. doi: 10.1007/s12374-015-0103-z
http://dx.doi.org/10.1007/s12374-015-0103-z
T. P. Sun , & F. Gubler. (2004). Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol, 55(1), 197-223. doi: 10.1146/annurev.arplant.55.031903.141753
http://dx.doi.org/10.1146/annurev.arplant.55.031903.141753
L. Thiery, A. S. Leprince, D. Lefebvre, M. A. Ghars, et al. (2004). Phospholipase D is a negative regulator of proline biosynthesis in Arabidopsis thaliana. J Biol Chem, 279(15), 14812-14818. doi: 10.1074/jbc.M308456200
http://dx.doi.org/10.1074/jbc.M308456200
A. Vashisht , & N. Tuteja. (2006a). Stress responsive DEAD-box helicases: a new pathway to engineer plant stress tolerance. J Photochem Photobiol B, 84(2), 150-160. doi: 10.1016/j.jphotobiol.2006.02.010
http://dx.doi.org/10.1016/j.jphotobiol.2006.02.010
A. Vashisht , & N. Tuteja. (2006b). Stress responsive DEAD-box helicases: a new pathway to engineer plant stress tolerance. J Photochem Photobiol B, 84(2), 150-160. doi: 10.1016/j.jphotobiol.2006.02.010
http://dx.doi.org/10.1016/j.jphotobiol.2006.02.010
Chun-Mei Wang, Jin-Lin Zhang , Xue-Song Liu, Zhan Li, et al. (2009). Puccinellia tenuiflora maintains a low Na+ level under salinity by limiting unidirectional Na+ influx resulting in a high selectivity for K+ over Na+ Plant, Cell & Environment Volume 32, Issue 5. Plant, Cell & Environment, 32(5), 486-496.
http://dx.doi.org/10.1111/j.1365-3040.2009.01942.x
J. Wang, L. Chen, Y. Wang, J. Zhang, et al. (2013). A computational systems biology study for understanding salt tolerance mechanism in rice. PLoS One, 8(6), e64929. doi: 10.1371/journal.pone.0064929
http://dx.doi.org/10.1371/journal.pone.0064929
S. Wang, Y. Bai, C. Shen, Y. Wu, et al. (2010). Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct Integr Genomics, 10(4), 533-546. doi: 10.1007/s10142-010-0174-3
http://dx.doi.org/10.1007/s10142-010-0174-3
Y Wang, Y Chu, G. Liu, M. H. Wang, et al. (2007). Identification of expressed sequence tags in an alkali grass (Puccinellia tenuiflora) cDNA library. J Plant Physiol, 164(1), 78-89. doi: 10.1016/j.jplph.2005.12.006
http://dx.doi.org/10.1016/j.jplph.2005.12.006
Wei, R. Z. Zhang, J. J. Guo, D. M. Liu, et al. (2014). Genome-wide analysis of the MADS-box gene family in Brachypodium distachyon. PLoS One, 9(1), e84781. doi: 10.1371/journal.pone.0084781
http://dx.doi.org/10.1371/journal.pone.0084781
L. Xiong, K. S. Schumaker, & J. K. Zhu. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell, 14 l(Supplement), S165-183.
X.F Yan, G.R Sun , J.X Li , J.L Li , et al. (1994). Primary studies on salt excretion ability of Puccinellia tenuiflora. Pratacultural Science 11.
T. Zhang, Y. Liu, T. Yang, L. Zhang, et al. (2006). Diverse signals converge at MAPK cascades in plant. Plant Physiol Biochem, 44(5-6), 274-283. doi: 10.1016/j.plaphy.2006.06.004
http://dx.doi.org/10.1016/j.plaphy.2006.06.004
JK Zhu (2002). Salt and drought stress signal transduction in plants. Annu Rev Plant Biol, 53(4), 247-273. doi: 10.1146/annurev.arplant.53.091401.143329
http://dx.doi.org/10.1146/annurev.arplant.53.091401.143329
. PDF(1644KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Panpan Liu
. Yajun Sun
. Xiaoxue Ye
. James X.J. Fang
Related articles
. Puccinellia tenuiflora
. NaHCO 3 stress
. cDNA clone
. Function annotation
Tools
. Email to a friend
. Post a comment