2 College of Agronomy and Biotechnology, Yunnan Agricultural University, Kunming, 650201, P.R. China
 Author
Author  Correspondence author
Correspondence author
Bioscience Methods, 2022, Vol. 13, No. 2 doi: 10.5376/bm.2022.13.0002
Received: 13 Jan., 2022 Accepted: 20 Jan., 2022 Published: 21 Feb., 2022
Yang H., Chen J., Zhang C.L., Arshad K.T., Li Q.P., Wang T., Wen J.C., and Li D.D., 2022, Construction of rice OsSUTs gene knockout lines using CRISPR/Cas9 system, Bioscience Method, 13(2): 1-8 (doi: 10.5376/bm.2022.13.0002)
The efficiency of sucrose transfer directly affects starch accumulation in rice grains and rice yield. There are five sucrose transporters reported in rice, the relationships of them are complex, and the interaction mechanisms of them are still unclear. In this study, based on the sequence information of the five OsSUTs genes in rice, target sites were designed respectively, the corresponding CRISPR/Cas9 vector was successfully constructed, and five positive knock-out seedlings of OsSUTs genes were obtained through transgenic technology. It provides a good material for exploring the molecular function and regulatory relationship of OsSUTs gene.
Background
Sucrose transporter is an important regulatory molecule for the transport and distribution of sucrose in plants. The rational and effective distribution of carbohydrates is achieved through the coordinated transport of sucrose/H+ across the membrane (Lei et al., 2011). In plants, SUTs genes exist in the form of multigene family. Various SUTs genes coordinate the normal physiological metabolism, growth and development of plants through complementation or cotransport (Chang et al., 2019). Studies have shown that SUTs genes play an important role in plant stress responses such as high temperature, high salt, drought and heavy metals (Gong et al., 2013; Shinichiro et al., 2017; Gong et al., 2019).
As an important food crop in the world, the yield of rice has always been an important goal of cultivation and breeding (Yamaguchi et al., 2020). The efficiency of sucrose transfer directly affects starch accumulation in rice grains and rice yield (Li et al., 2019). There are 5 OsSUTs genes (OsSUT1~OsSUT5) reported in rice (Aoki et al., 2003), and OsSUTs genes regulate sucrose transport rate through different spatio-temporal expression patterns, affecting plant growth and reproductive development (Feng et al., 2018; Yue, 2020). In different growth periods and organs and tissues, OsSUTs genes are overlapped and differentially expressed, which means that there are complex regulatory relationships of coordination and complementation among members (Eom et al., 2016). However, few studies have been reported on the interaction between members of the OsSUTs gene family. Therefore, it is of great significance to explain the yield of rice by exploring the interaction between five OsSUTs genes and clarifying the intermolecular regulation mechanism of genes.
As a new gene editing technology in recent years, CRISPR/Cas9 system is an accurate and efficient technology to explore the biological function and molecular interaction mechanism of gene molecules. It combines RNA (gRNA) with gene target sequence, splits the target DNA molecule under the action of Cas9 protein, and automatically repairs DNA molecule after breakage. And base insertion/deletion or substitution occurs in the process of repair, resulting in early termination of translation and the loss of the function of the original gene (Wang et al., 2018; Shen et al., 2019). In plants, there are many gene knockout models constructed by this technology, such as the gene knockout model of catalase (CAT) in rice. Since the function of CatB involved in abiotic stress response is largely unknown, this knockout model is constructed to analyze the physiological and biochemical phenotypes of catb under salt, high temperature and oxidation stresses (Liu et al., 2020). The gene knockout model of fat dehydrogenase AhFAD2 in peanut was constructed to research the function of AhFAD2 gene. The omega-6 fatty acid dehydrogenase controls the conversion of oleic acid into linoleic acid in peanut, which is encoded by the AhFAD2 gene (Li et al., 2019). In order to explore the function and regulatory mechanism of CPS2 gene, the gene knockout model of CPS2 in Nicotiana tabacum is constructed (Zhang et al., 2020). And therefore, it laid an important foundation for exploring the molecular function and regulation pathway of genes, as well as crop genetic improvement and new variety cultivation.
In this study, based on the sequence information of the five OsSUTs genes in rice, target sites were designed respectively, the corresponding CRISPR/Cas9 vector was successfully constructed, and five positive knock-out seedlings of OsSUTs genes were obtained through transgenic technology. It provides a good material for exploring the molecular function and regulatory relationship of OsSUTs gene.
1 Results and Analysis
1.1 Design of target sites for OsSUTs gene knockout
The DNA sequences of 5 OsSUTs genes were searched in NCBI database, and the corresponding gene structure was analyzed according to the CDS region of each gene, and the conserved region of the transmembrane domain of the gene was identified. The target sites of OsSUTs gene knockout were designed by Guide Design Resources website (http://crispr.mit.edu). In order to improve the knockout efficiency, two targets were designed for each gene based on the strategy of constructing double-target vectors. The target sites of OsSUT1 gene were located in exon 9 and exon 11, those of OsSUT2 and OsSUT3 were located in exon 1, OsSUT4 was in exon 1 and exon 14, and OsSUT5 was in exon 1 and exon 2 (Figure 1).
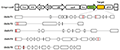 Figure 1 Structure and target location of the OsSUTs knockout vectors |
1.2 Construction of OsSUTs gene knockout vectors
The CRISPR-Cas9 vector was digested with restriction endonuclease Bsa Ⅰ and Eco31 Ⅰ, and the products were separated by electrophoresis. The vector fragment of about 13 000 bp was cut off and purified. The 5 target sites with restriction sites were digested with the above two endonucleases, and the digested products were purified. T4 ligase was used to connect the target site sequence with the recovered product after CRISPR-Cas9 digestion. Cas9::SUT1, Cas9::SUT2, Cas9::SUT3, Cas9::SUT4, Cas9::SUT vectors were constructed, respectively. The ligated vector was transformed into E. coli, and monoclonal clones were selected for PCR identification. The positive clones were 714 bp (OsSUT1), 399 bp (OsSUT2), 315 bp (OsSUT3), 515 bp (OsSUT4), and 1 228 bp (OsSUT5), respectively (Figure 2A). The 5 constructed vectors were verified by restriction digestion (Figure 2B), and finally the vectors were transformed into rice callus with Agrobacterium-mediated genetic transformation.
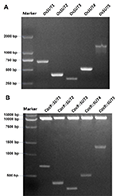 Figure 2 Electrophoresis of OsSUTs gene target region and restriction digestion verification of knockout vector Note: A: Gel electrophoresis of the PCR productions of target regions for knocking-out OsSUTs genes; B: Enzyme cutting bands; Marker: Trans15K DNA Marker |
1.3 Detection of positive knock-out seedlings of rice OsSUTs genes
The transformed rice callus was screened by Kana. After inducing germination and rooting, it was transferred to the greenhouse for trained transition and transplantation, and 234 transgenic rice seedlings of T0 generation were obtained. Genotype identification (Figure 3A) and DNA sequence alignment (Figure 3B) were performed on the target regions of OsSUTs gene knockout in these transgenic plants. The results showed that among the 42 OsSUT1 gene knockout transgenic plants, 3 PCR products had overlapping peaks, and the sequence had low homology with OsSUTs reported sequence, which were positive knock-out seedlings, accounting for 7.14% of the transgenic seedlings. Among the 50 OsSUT2 gene knockout transgenic plants, 7 were positive, accounting for 14.0%. Among the 50 OsSUT3 gene knockout transgenic plants, 17 were positive, accounting for 34.0%. Among 42 OsSUT4 gene knockout transgenic plants, 4 were positive, accounting for 9.52%. Among 50 OsSUT5 gene knockout transgenic plants, 16 were positive, accounting for 32.00%. In which, the knockout rates of OsSUT3 and OsSUT5 were the highest, both above 30.0%, while the OsSUT1 and OsSUT4 were the lowest, both below 10.0%.
 Figure 3 Sequencing of target regions in positive transgenic rice for knocking-out OsSUTs Note: A: Electrophoresis and sequencing peak maps of PCR amplified products from the target region of OsSUTs gene knockout transgenic rice seedlings; B: Sequence alignment of OsSUTs target gene and knockout positive vaccine target region |
2 Discussion
In plants, SUTs proteins are relatively conserved in the process of evolution, with highly homologous amino acid sequences and similar structures, all of which have 12 transmembrane domains (Sun, 2012). Therefore, SUTs proteins have similar biological functions in plants, which makes it difficult to study the functional differences among members of the SUTs family. According to previous studies, the main function of rice OsSUTs protein is to coordinate the transport and reasonable distribution of carbon in plants, but the different spatio-temporal expression between each member shows that there are also significant functional differences among OsSUTs members at different stages of rice growth and development (Feng, 2018; Zhang et al., 2014). There are many studies on OsSUT1 gene. OsSUT1gene can regulate rice early germination, seedling development and grain filling (Kato and Horibata., 2011; Feng et al., 2018), but also can coordinate the stress response of plants under high temperature and high salt environment (Siahpoosh et al., 2012), indicating that OsSUT1 can accelerate the transport rate of sucrose in plants during the vigorous growth of rice and provide sufficient energy for the physiological metabolism of rice (Scofield et al., 2007). OsSUT2, located on vacuole membrane, is highly expressed in many tissues and organs, such as rice mesophyll, lateral root, pedicel after fertilization and so on, mainly coordinating the storage and transport of sugar molecules in vacuoles (Siao et al., 2011). This gene mutation will lead to changes in important agronomic characters of rice, such as the decrease of tiller number, plant height and 1000-grain weight (Yan et al., 2011; Du, 2010; Hong, 2008). However, maybe because of the high homology of OsSUTs gene sequence, there are relatively few studies on OsSUT3, OsSUT4 and OsSUT5 genes (Zhang et al., 2014). Previous studies carried out site-directed mutagenesis on OsSUT1 and OsSUT3 genes to study the effects of these two genes on rice pollen development. However, only OsSUT1 mutants were obtained, while OsSUT3 mutants were not obtained (Hirose et al., 2010). It is further indicated that the construction of rice OsSUTs knockout lines is of great significance to further explore the biological functions of OsSUTs members and their effects on rice agronomic traits.
In recent years, with the gradual maturity of CRISPR/Cas9 technology, only one or two target sequences can be designed and synthesized to guide Cas9 protein to specifically bind and modify the target gene, leading to gene mutation or knockout. Homozygous plants can be obtained in the T0 generation. It is widely used in plant genetic breeding (Khlestkina and Shumny, 2016), and greatly promotes the development of gene basic research (Meiliana et al., 2017). In this study, 5 rice OsSUTs gene knockout vectors were constructed by CRISPR/Cas9 technology, and 5 OsSUTs gene knockout lines were obtained by genetic transformation. However, after molecular screening analysis, it was found that the proportion of OsSUTs gene knockout in these 5 vectors was significantly different. Among them, OsSUT3 and OsSUT5 had the highest proportion of knockout, followed by OsSUT2, and OsSUT4 and OsSUT1 had the lowest proportion of knockout. It is speculated that the target sites of OsSUT2, OsSUT3 and OsSUT5 are located on the exon of the 5' end of the gene, so the binding of the target can inhibit the transcriptional translation of the gene as soon as possible. While the target site of OsSUT1 or OsSUT4 is located on the 3' end of the gene, so that the binding of the target does not change the sequence of the 5' end of the gene, which has relatively little effect on gene transcription and translation, resulting in a significant reduction in the proportion of gene knockout. Therefore, the design of target sequences is very important when using CRISPR/Cas9 system for gene editing. This is also confirmed in previous studies. For the target designed for mouse mmu-miR-155 seed sequence, the target designed by Jing et al. (2015) knocked out at most 6 bp, and the mRNA expression level of mmu-miR-155 was only reduced by half, while the target designed by Li et al. (2019) knocked out 31 bp, and the mRNA expression level of mmu-miR-155 was directly reduced by more than 8~10 times.
In summary, in this study, 5 rice OsSUTs gene knockout lines were successfully constructed by CRISPR/Cas9 system, which provided good material for exploring the molecular function and regulatory relationship of OsSUTs gene, and laid a solid foundation.
3 Materials and Methods
3.1 Test materials and reagents
Materials Nipponbare, E. coli DH5α and Agrobacterium GV3101 were provided by Rice Farming Research Institute, Yunnan Agricultural University. CRISPR-Cas9 gene editing vector was purchased from Wuhan BioRun Co., Ltd.
Restriction endonuclease Bsa Ⅰ and Eco31 Ⅰ were purchased from New England Biolabs, T4 ligase was purchased from TaKaRa, Gel Extraction Kit and Plasmid DNA Extraction Kit were purchased from TIANGEN, synthesis and sequencing of primers and targets were completed by Sangon Biotech (Shanghai) Co., Ltd. (Table 1).
 Table 1 The sequences of targets and primers used in this study Note: Underlined letters: The sites recognized by Bsa Ⅰ and Eco31 Ⅰ |
3.2 Agrobacterium-mediated transformation
According to the method of Saha et al. (2011), the constructed vector plasmid was transformed into Agrobacterium under the condition of electric shock at 2 500 V/6 ms, and the positive transformation strains were screened to culture for further use. The seeds of Nipponbare were disinfected with 0.1% HgCl2 and inoculated on the induction medium for callus culture. The rice callus was infected with the cultured activated Agrobacterium, and then the infected callus was transferred into the screening medium, germination medium and rooting medium, respectively. The transformed seedlings were cultivated under 16 h light and 8 h darkness, and transplanted after 15 d of trained transition in greenhouse.
3.3 Target sequence detection of target genes in transformed plants
The modified 2×CTAB method (Doyle and Doyle, 1987) was used to extract DNA from the seedling leaves of the transformed plants, and the specific primers were used for PCR amplification. The amplified products were detected by agarose gel electrophoresis. The purified products were sequenced and compared, and the positive knockout gene plants were screened.
Authors’ contributions
YH and CJ were the experimental designers and executor of this study. YH and LDD completed the data analysis and the writing of the first draft of the paper. ZCL, AKT, LQP, WT participated in the experimental design and analysis of the experimental results. LDD and WJC were responsible for guiding the experimental design, data analysis and manuscript writing and revision. All authors read and approved the final manuscript.
Acknowledgments
This study was supported by the Project of Yunnan Academician Workstation (2018IC065) and National Natural Science Foundation of China (32060137).
Aoki N., Hirose T., Scofield G.N., Whitfeld P.R., and Furbank R.T., 2003, The sucrose transporter gene family in rice, Plant Cell Physiol., 44(3): 223-232
https://doi.org/10.1093/pcp/pcg030
PMid:12668768
Chang Y.A., Dai N.C., Chen H.J., Tseng C.H., and Wang S.J., 2019, Regulation of rice sucrose transporter 4 gene expression in response to insect herbivore chewing, J. Plant Interact., 14(1): 525-532
https://doi.org/10.1080/17429145.2019.1662099
Doyle J., and Doyle J.J., 1987, A rapid DNA isolation procedure for small quantities of fresh leaf tissue, Phytochem. Bull, 19(1): 11-15
Du L., 2010, Molecular regulation of OsSUT on rice grout physiology, Thesis for M.S., Fujian Agriculture and Forestry University, Supervisors: Zheng J.G., and Su J., pp.35-40
Eom J.S., Nguyen C.D., Lee D.W., Lee S.K., and Jeon J.S., 2016, Genetic complementation analysis of rice sucrose transporter genes in arabidopsis SUC2 mutant atsuc2, J. Plant Biol., 59(3): 231-237
https://doi.org/10.1007/s12374-016-0015-6
Feng B., 2018, Role of rice sucrose transporter gene OsSUT1 in rice growth and development and phosphorus nutrition, Thesis for M.S., Nanjing Agricultural University, Supervisor: Sun S.B., pp.50-55
Feng B., Sun Y.F., Ai H., Liu X.L., Yang J., Liu L., Gao F.Y., Xu G.H., and Sun S.B., 2018, Overexpressed of sucrose transporter OsSUT1 affects rice morphology and physiology, Zhongguo Shuidao Kexue (Chinese Science of Rice), 32(6): 549-556
Gong X., Jiang M., Qi X., Liu X.F., Lu S.Q., Chen K., Liu Y.L., Zhang S.K., Ma T., and Yang Y.J., 2019, Cloning of sucrose transporter gene ZmSUT4 from maize and its expression analysis under low temperature stress, Nanfang Nongye Xuebao (Journal of Southern Agriculture), 50(6): 1165-1172
Gong X., Liu M., Wang C., Zhang L., and Liu W., 2013, Sucrose transporter gene AtSUC4 regulates sucrose distribution and metabolism in response to salt stress in Arabidopsis thaliana, Advanced Materials Research, 56(8): 1574-1587
https://doi.org/10.4028/www.scientific.net/AMR.726-731.217
Hirose T., Zhang Z.J., Miyao A., Hirochika H., Ohsugi R., and Terao T., 2010, Disruption of a gene for rice sucrose transporter, OsSUT1, impairs pollen function but pollen maturation is unaffected, J. Exp. Bot., 61(13): 3639-3646 3639-3646
https://doi.org/10.1093/jxb/erq175
PMid:20603282 PMCid:PMC2921200
Hong H.Q., 2008, Over expressing the grout physiology of OsSUT2 and OsSUT5 indica rice, Fujian Agriculture and Forestry University, Supervisor: Wang F., pp.35-40
Jing W.X., Zhang X.W., Sun W.Y., Hou X.J., Yao Z.Q., and Zhu Y.L., 2015, CRISPR/CAS9-mediated genome editing of miRNA-155 inhibits proinflammatory cytokine production by RAW264.7 cells, Biomed Research International, 2015: 1-7
https://doi.org/10.1155/2015/326042
PMid:26697483 PMCid:PMC4677169
Kato T., and Horibata A., 2011, Non-random distribution of the alleles for good grain filling at OsAGPS2 and OsSUT1 among a wide range of rice (Oryza sativa L.) cultivars, Breeding Science, 61(2): 217-220
https://doi.org/10.1270/jsbbs.61.217
Khlestkina E.K., and Shumny V.K., 2016, Prospects for application of breakthrough technologies in breeding: The CRISPR/Cas9 system for plant genome editing, Russian Journal of Genetics, 52(7): 676-687
https://doi.org/10.1134/S102279541607005X
Lei M., Liu Y.D., Zhang B.C., Zhao Y.T., Wang X.J., Zhou Y.H., Raghothama K.G., and Liu D., 2011, Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in arabidopsis, Plant Physiol., 156(3): 1116-1130
https://doi.org/10.1104/pp.110.171736
PMid:21346170 PMCid:PMC3135933
Li C.C., Zhang Y.H., Zhao W.X., Wu J., Han H.Y., Li M.Y., Niu H., Song S.F., and Li W.T., 2019, Preparation of Mir-155 gene knockout cells mediated by CRISPR/Cas9 system, Shengwu Jishu Tongbao (Bulletin of Biotechnology), 35(11): 231-239
Li G.H., Zhou C.Y., Guo B.W., Wei H.Y., Huo Z.Y., Dai Q.G., Zhang H.C., and Xu K., 2019, Sucrose phloem loading and its relationship with yield formation in rice, Zhiwu Shengli Xuebao (Journal of Plant Physiology), 55(7): 891-901
Li K., Zhao K.K., Ning L.L., Ma Q., Li Z.F., Ma X.L., Zhang X.G., and Yin D.M., 2019, Multi-target knockout vector construction of peanut FAD2 gene CRISPR/Cas9, Shandong Nongye Kexue (Shandong Agricultural Science), 51(9): 56-67
Liu S., Zhao L.J., Liu C., Deng Y., Huang J., Tang D.Y., Liu X.M., and Lin J.Z., 2020, Construction of rice OsCatB knockout mutant and preliminary analysis of their stress tolerance, Shengmin Kexue Yanjiu (Life Sciences Research), 24(4): 301-309
Meiliana A., Dewi N.M., and Wijaya1 A., 2017, Genome editing with Crispr/Cas9 systems: basic research and clinical applications, The Indones. Biomed. J., 9(1): 1-16
https://doi.org/10.18585/inabj.v9i1.272
Saha D., Kumar V., Bhat S.R., and Srinivasan R., 2011, Characterization of upstream sequences of the loj gene leads to identification of a novel enhancer element conferring lateral organ junction-specific expression in Arabidopsis thaliana, Plant Mol. Biol. Rep., 29(2): 265-277
https://doi.org/10.1007/s11105-010-0229-6
Scofield G.N., Aoki N., Hirose T., Takano M., Colin L.D., Jenkins., and Furbank R.T., 2007, The role of the sucrose transporter, OsSUT1, in germination and early seedling growth and development of rice plants, J. Exp. Bot., 58(3): 483-495
https://doi.org/10.1093/jxb/erl217
PMid:17138625
Shen L., Hua Y., Fu Y., Li J., Liu Q., Jiao X., Xin G., Wang J., Wang X., Yan C., and Wang K., 2019, Erratum to: rapid generation of genetic diversity by multiplex Crispr/Cas9 genome editing in rice, Science China Life Sciences, 62(9): 506-515
https://doi.org/10.1007/s11427-019-9599-5
PMid:31407141
Shinichiro T., Ayano M.M., and Midori Y., 2017, Analysis of sugar content and expression of sucrose transporter genes (OsSUTs) in rice tissues in response to a chilling temperature, Japan Agricultural Research Quarterly: JARQ, 51(2): 137-146
https://doi.org/10.6090/jarq.51.137
Siahpoosh M.R., Sanchez D.H., Schlereth A., Scofield G.N., Furbank R.T., Dongen J.T.V., and Kopka J., 2012, Modification of OsSUT1 gene expression modulates the salt response of rice Oryza sativa cv. Taipei 309, Plant Sci., 182: 101-111
https://doi.org/10.1016/j.plantsci.2011.01.001
PMid:22118621
Siao W., Chen J.Y., Hsiao H.H., Chung P., and Wang S.J., 2011, Characterization of OsSUT2 expression and regulation in germinating embryos of rice seeds, Rice, 4(2): 39-49
https://doi.org/10.1007/s12284-011-9063-1
Sun Y, 2012, Structure-function relationship of plant sucrose transporters (SUTS), Dissertations for Ph.D., University of Minnesota, Supervisor: Ward J.M., pp.2-6
Wang P., Zhang J., Sun L., Ma Y., and Zhang X., 2018, High efficient multisites genome editing in allotetraploid cotton (gossypium hirsutum) using Crispr/cas9 system, Plant Biotech. J., 16(1): 137-150
https://doi.org/10.1111/pbi.12755
PMid:28499063 PMCid:PMC5785356
Yamaguchi K., Yamamoto T., Segami S., Horikawa M., & Miura K., 2020, Gw2 mutation increases grain width and culm thickness in rice (Oryza sativa L.), Breeding ence, 70(4): 456-461
https://doi.org/10.1270/jsbbs.20018
PMid:32968348 PMCid:PMC7495194
Yan C.J., Tian Z.X., Fang Y.W., Yang Y.C., Li J., Zeng S.Y., Gu S.L., Xu C.W., Tang S.Z., and Gu M.H., 2011, Genetic analysis of starch paste viscosity parameters in glutinous rice (Oryza sativa L.), Theor. Appl. Genet., 122(1): 63-76
https://doi.org/10.1007/s00122-010-1423-5
PMid:20737264
Yue M.M., 2020, Functional analysis of rice sucrose transporter OsSUT4, Dissertation for Ph.D., China Agricultural University, Supervisors: Meng Q.W. and Lu C.M., pp.1-10
Zhang S.Q., He J., He L.X., Xue G., Sun J.T., Li X.H., Du W.D., and Xu S.X., 2020, Gene knockout and functional analysis of CPS2 in tobacco based on CRISPR/Cas9 technology, Yancao Keji (Tobacco Science and Technology), 53(5): 17-25
Zhang W.J., Guan Q.L., Fu Y.P., and Su J., 2014, Antisense suppression expression of rice (Oryza sativa L.) sucrose transporter gene (OsSUT5) leads to reducing callus induction and plantlet regeneration, Nongye Shengwu Jishu Xuebao (Journal of Agricultural Biotechnology), 22(7): 825-825
. PDF(1006KB)
. HTML
Associated material
. Readers' comments
Other articles by authors
. Hong Yang
. Jin Chen
. Chunlong Zhang
. Khadija Tehseen Arshad
. Qiuping Li
. Tuo Wang
. Jiancheng Wen
. Dandan Li
Related articles
. Rice ( Oryza sativa L.)
. OsSUT genes
. Gene knockout
. CRISPR/Cas9
Tools
. Email to a friend
. Post a comment


