Research Article
Genome-wide Identification and Expression Analyses of PAL Genes in Different Color Radish 
2 Fujian EcoTech Center for State Farm and South Subtropical Crops, Fuzhou, 350003, P.R. China
 Author
Author  Correspondence author
Correspondence author
Computational Molecular Biology, 2022, Vol. 12, No. 1 doi: 10.5376/cmb.2022.12.0001
Received: 22 Dec., 2021 Accepted: 14 Jan., 2022 Published: 28 Feb., 2022
Lai B., Chen C.F., Tang Z.H., Xiao J., Wang Q., Chen F.B., and Du L.N., 2022, Genome-wide identification and expression analyses of PAL genes in different color radish, Computational Molecular Biology, 12(1): 1-9 (doi: 10.5376/cmb.2022.12.0001)
Phenylalanine ammonia-lyase (PAL) is the first step key enzyme of the flavonoid biosynthesis pathway which played an essential role in plant anthocyanin accumulation. Five PAL family gene members were identified in the radish (Raphanus sativus L.) genome name RsPAL1~5. Specific primers were designed to amplify the open reading frame in red radish ‘Hongxin No.1’ and then sequenced. Real-time PCR was used to analysis the expression pattern of 5 RsPALs in four different tissues including leaf, petiole, taproot flesh and skin form five different color type radishes, including red skin and red flesh ‘Shaguan’ and ‘Hongxin No.1’, red skin and white flesh ‘Shaguan No.1’ and ‘Mantanghong’, white skin and white flesh ‘Chunbulao’. The results showed that the length of open read frame of RsPAL1~5 were 2 160, 2 166, 2 163, 2 124 and 2 109 bp encoding for 719, 721, 720, 707 and 702 amino acid residues respectively. Sequence alignment analysis showed that MIO motif (Ala-Gly-Ser) was conserved among the five RsPALs proteins. RsPAL1~4 were clustered with Arabidopsis AtPAL1 and AtPAL2, RsPAL5 was clustered with AtPAL4 in the phylogenetic tree. Real-time PCR results suggested that RsPAL4 was expressed only in the tissue accumulate anthocyanin, and the expression of RsPAL4 was significantly correlated with anthocyanin content. These results indicating that RsPAL4 may specifically involved in anthocyanin biosynthesis in radish. However, no obvious expression pattern of other RsPALs members was found in this study, suggesting that they may participate in other secondary metabolites biosynthesis of phenylpropane metabolism pathway. This study would provide scientific basis for further study on the function of radish PAL.
Background
Carmine radish (Raphanus sativus), also known as ‘Hongxin’ radish, is a local variety in the genus Raphanus of Brassicaceae. It is mainly abundant in Chongqing and Sichuan. Due to the accumulation of a large amount of anthocyanin in its fleshy roots, it presents dark red color from epidermis to interior. In recent ten years, the value of anthocyanin has been recognized, such as anti-oxidation, anti-inflammatory, anti-aging, and anti-cancer effects, as well as liver, cerebrovascular and vision protective effects (Xu et al., 2013). Anthocyanin is an important secondary metabolite in the flavonoid family and an important pigment in plant organs from red to purple. Its biosynthetic metabolic pathway has been clearly studied (Winkel-Shirley, 2001). PAL is the first step key enzyme and rate-limiting enzyme of the phenylpropane metabolism pathway, which is mainly responsible for catalyzing the deamination of L-Phenylalanine to trans-Cinnamic acid (Koukol et al., 1961). In addition to flavonoids, trans-Cinnamic acid is also involved in the synthesis of lignin and phytoalexin and other secondary metabolites. Therefore, PAL plays an essential role in plant growth and development and stress resistance (Hao et al., 2018).
PAL genes in plants are generally composed of several family members. 4, 6 and 5 PAL family members were found in Cucumis melo, Citrullus lanatus, and Cucumis sativus, respectively (Sun et al., 2018). 9 members of the OsPALs family were also identified in Oryza sativa, and it was found that 8 of them were involved in the response to abiotic stress (Zeng et al., 2018). 8 members of MaPALs gene family were found in Musa nana. The expression of different members in different stages of fruit development and abiotic stress was significantly different, indicating that the biological functions of different members were also different (Yang et al., 2019). In addition, members of the PAL family were identified from many horticultural plants, such as Vitis amurensis (Chen et al., 2018), Punica granatum (Feng et al., 2018), Solanum tuberosum (Chang et al., 2018), and Luffa cylindrical (Zhu et al., 2018).
Early studies have found that the synthesis of apple anthocyanin was closely related to PAL enzyme activity. The higher PAL enzyme activity, the better apple colouring (Zhou et al., 1997). PAL activity in the peel of Camellia oleifera was not only positively correlated with its resistance to Colletotrichum gloeosporioides, but also positively correlated with anthocyanin content in the peel (Yang et al., 2007). The accumulation of anthocyanin in radish seedlings was induced by UV treatment, and the PAL activity was also increased. And it was found that PAL activity was also positively correlated with anthocyanin content in different tissues and different developmental stages (Su et al., 2015; Zhang et al., 2019). However, there is no specific RsPAL family member involved in anthocyanin biosynthesis at present in radish at the whole gene level, although it has been reported that some RsPAL genes are not related to anthocyanin biosynthesis (Muleke et al., 2017).
In this study, RsPALs gene family members were screened from RadishBase and their structures were analyzed. Using carmine radish 'Hongxin No.1' as material, the RsPALs gene was amplified and sequenced, and the expression of RsPALs members in different tissues of different color radish varieties was detected by Real-time fluorescence quantitative PCR. The correlation between the expression level of RsPALs gene and anthocyanin content was analyzed, and the key RsPALs gene involved in anthocyanin synthesis were identified, which provided a theoretical basis for radish variety improvement.
1 Results and Analysis
1.1 Anthocyanin content analysis in different color radish
In this study, the anthocyanin contents in leaf, petiole, skin and flesh of different radish varieties were determined by pH-differential method (Figure 1). The fleshy roots of 'Shaguan' and 'Hongxin No.1' were red skin and red flesh, and the petioles were red, and there were a lot of anthocyanin accumulation. Among them, the average anthocyanin content in the skin of 'Shaguan' was 1.95 mg/g. The fleshy roots of red skin and white flesh 'Mantanghong' and 'Shaguan No.1' were red skin and white flesh, and the petioles were red. There were a lot of anthocyanins in the taproot flesh and petioles of 'Shaguan No.1'. While the content of 'Mantanghong' was less. Anthocyanin accumulation was not detected in petiole, taproot flesh and flesh in ‘Chunbulao’. And no anthocyanin was found in the leaves of 5 radish varieties.
|
Figure 1 Anthocyanin content in different tissues from different radish cultivars |
1.2 Identification and cloning of PAL family members in radish
According to the 4 reported AtPAL protein sequences of Arabidopsis thaliana, 5 members of radish RsPALs family with high homology were searched by Blastp software from RadishBase and further confirmed by BlastCCD analysis. The gene numbers were RSG04490.t1, RSG09128.t1, RSG33787.t1, RSG39829.t1 and RSG12055.t1, respectively, and named RsPAL1~5. cDNA of carmine radish ‘Hongxin No.1’ was used as the template, specific primers were designed to amplify and sequence the 5 genes, and open reading frames of RsPALs members in red radish were obtained, with the nucleotide sequences of 2 160, 2 166, 2 163, 2 124 and 2 109 bp, respectively. The results of sequence alignment showed that the similarity of amino acid sequences of RsPAL1 and RsPAL2 was the highest (95.29%), followed by that of RsPAL3 and RsPAL4 (94.48%). In addition, the similarity of amino acid sequences between RsPAL1 and AtPAL1 was 93.93%. The similarity of amino acid sequences between RsPAL1 and AtPAL3, RsPAL4 was relatively low, which was 70.66% and 79.14%, respectively. The similarity of amino acid sequences between RsPAL5 and AtPAL4 was 90.54% (Figure 2). Sequence alignment results showed that the amino acid sequences of carmine radish RsPAL family members were higher than those of Arabidopsis thaliana AtPAL family members, indicating that 5 carmine radish RsPALs family members were successfully cloned. Multiple sequence alignment analysis showed that the PAL active site GTITASGDLV(L)PLSYIAG was found in all amino acid sequences of PAL gene members in Raphanus sativus and Arabidopsis thaliana, which contained highly conserved MIO electrophilic group composed of Ala-Ser-Gly (Figure 2).
|
Figure 2 Amino acids sequences alignment of PAL family members from Arabidopsis and radish Note: In the red box is the active site of PAL protein; The red triangle showed the MIO domain |
1.3 Gene structure, protein structure and phylogenetic analysis of PAL family in carmine radish
TBtools software was used to analyze the exons and introns of 5 RsPALs family members according to genome annotation information. It was found that RsPAL1, RsPAL3 and RsPAL4 had only 1 intron and 2 exons, while RsPAL2 had 3 introns and 4 exons, and RsPAL5 had 2 introns and 3 exons. By using MEME online software to analyze the domains of the 5 RsPALs proteins, it was found that all the 5 members had the same 10 protein domains (Motif), and the structures of the 5 RsPALs proteins were generally consistent, indicating that the sequences among PAL members were conservative (Figure 3).
|
Figure 3 Gene structure and conserved protein motifs analysis of radish RsPALs family members Note: A: RsPAL exon-intron analysis; B: Conserved protein motif predicted by MEME software; C: Sequences of 10 conserved motifs |
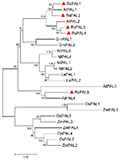
Neighbor-joining (NJ) phylogenetic tree was constructed to study the evolutionary relationship between RsPAL and PAL from other plants (Figure 4). It can be seen that PAL proteins from monocotyledonous Zea mays and Oryza sativa were clustered together, while PAL proteins from dicotyledonous plants Raphanus sativus, Glycine max, Arabidopsis thaliana, Lycopersicon esculentum and Nicotiana tabacum were clustered together (Figure 4). Among them, RsPAL3 and RsPAL4 were clustered with Arabidopsis thaliana AtPAL2, RsPAL1 and RsPAL2 were clustered with Arabidopsis thaliana AtPAL1, RsPAL5 and AtPAL4 were clustered with AtPAL5.
|
Figure 4 Neighbor joining phylogenetic tree of PAL from radish and other plants Note: Red triangle reparent PAL from radish |
1.4 Expression analysis of RsPAL in different color radish
To analyze the expression levels of these 5 members in the skin, flesh, petiole, and leaf in different color radish, including red skin and red flesh ‘Shaguan’ and ‘Hongxin No.1’, red skin and white flesh ‘Mantanghong’ and ‘Shaguan No.1’, and white skin and white flesh ‘Chunbulao’. It can be seen that RsPAL1 has the highest expression level in the taproot flesh of different radish varieties, and also has a high expression level in the petiole (Figure 5). RsPAL2 was highly expressed in the skin of ‘Shaguan’ and in the flesh of ‘Hongxin No.1’. RsPAL3 was highly expressed in the skin and flesh of different varieties, and in the petiole of ‘Hongxin No.1’. RsPAL5 was highly expressed only in the flesh and petiole of ‘Shaguan’. RsPAL4 was only highly expressed in tissues with anthocyanin accumulation. The higher the anthocyanin content, the higher the expression of RsPAL4, indicating that RsPAL4 is closely related to anthocyanin synthesis. It was found that the expression level of RsPAL4 in different tissues of different varieties was positively correlated with the content of anthocyanins, and the correlation coefficient was R2=0.794. In addition, the expression of RsPAL1/2/3/5 in different tissues of different varieties was not significantly correlated with anthocyanin content.
|
Figure 5 Expression of RsPALs in different tissues from different cultivars and the correlation between the expression of RsPAL4 and anthocyanin content Note: A~E: Expression of RsPAL1~RsPAL5 in in different tissues from different cultivars; F: Correlation between expression of RsPAL4 and anthocyanin content; The vertical bars represent the standard error of triplicate experiments |
2 Discussion
PAL is the first step key enzyme and rate-limiting enzyme of the phenylpropane metabolism pathway, which is a necessary pathway of secondary metabolism, affecting plant growth and development and response to stress. In this study, 5 RsPALs family members were identified in radish genome, and 1 more member than in Arabidopsis thaliana (Huang et al., 2010). Sequence alignment analysis showed that MIO motif (Ala-Gly-Ser) was conserved among the 5 RsPALs proteins. However, there were significant differences in protein conserved domains among the 8 MdPALs members in apple (Zhang et al., 2018). In evolution, RsPAL1 and RsPAL2 were clustered with Arabidopsis AtPAL1, RsPAL3 and RsPAL4 were clustered with Arabidopsis AtPAL2, and RsPAL5 was closely related to AtPAL3 and AtPAL4. In Arabidopsis thaliana, AtPAL4 is mainly involved in lignin synthesis, but its specific function is not clear because there is no phenotypic change after pal3 mutation alone. AtPAL1 and AtPAL2 were mainly involved in flavonoid biosynthesis, and the Arabidopsis anthocyanin biosynthesis were inhibited after pal1 and pal2 double mutations (Olsen et al., 2008; Huang et al., 2010).
A PAL member with a significant negative correlation with anthocyanin content was found in pomegranate (Punica granatum), suggesting that it may be involved in the browning process of pomegranate (Feng et al., 2018). In strawberry 'Camarosa', FaPAL6 gene expression was closely related to anthocyanin accumulation (Pombo et al., 2011). In this study, the expression levels of 5 RsPALs family members in different tissues of different color radish were analyzed. The results showed that RsPAL4 gene was expressed in the tissues of anthocyanin accumulation and was positively correlated with anthocyanin content, with the correlation coefficient of R2=0.794, indicating that RsPAL4 was specifically involved in radish anthocyanin biosynthesis, which provided gene resources for radish breeding with high anthocyanin content.
RsPAL1~3 and RsPAL5 did not show obvious regularity in the samples of this experiment, which may be due to their participation in other secondary metabolic pathways of radish. For example, RsPAL1 was relatively high expressed in the skin of different radish, suggesting that it may be involved in lignin synthesis. Because in general, the texture of radish skin is harder, and the content of lignin is higher (Li et al., 2008). 13 members of PAL gene family were identified in maize, 10 of which could be induced by sheath-blight fungus, and most of them were involved in the process of disease resistance (Deng et al., 2019). In addition, under various biological stress treatments, the expression of 6 PAL family members in Camellia sinensis was significantly increased (Xiong et al., 2020). Therefore, further studies are needed to better understand the specific secondary metabolic pathways involved in these members, thereby affecting radish growth and stress response.
3 Materials and Methods
3.1 Experimental materials
Different color radish used in this study include red skin and red flesh 'Shaguan' and 'Hongxin No.1', red skin and white flesh 'Shaguan No.1' and 'Mantanghong', and white skin and white flesh 'Chunbulao'. All experimental materials were planted at the experimental station of Yangtze Normal University (29°45′E, 107°15′N) in July 2016, and samples were collected in February 2017. The leaf, petiole, taproot flesh and flesh of different varieties were separated and cut into small pieces, and then immediately frozen in liquid nitrogen and stored in refrigerator (-80°C) for further use.
3.2 Determination of anthocyanin content
pH-differential method was used to determine the anthocyanin content according to the method of Wrolstad et al. (1982).
3.3 Identification of RsPAL family members
Amino acid sequences of 4 reported AtPALs members of Arabidopsis thaliana (AtPAL1, AT2G37040; AtPAL2, AT3G53260; AtPAL3, AT5G04230; AtPAL4, AT3G10340) were searched by Blastp software from RadishBase (http://www.nodai-genome-d.org), and then the NCBI Blast CD-search tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) was used to detect whether the amino acid sequences speculated by the candidate genes had a typical PAL protein domain (PF00221). The conserved protein domain analysis of RsPALs members was completed by the online tool MEME (http://meme-suite.org/tools/meme). The maximum number of conserved domains was set to 10, and the other parameters were the default parameters. The gene structure of RsPALs members was drawn according to the annotation information of radish genome by tbtools software (Chen et al., 2018).
3.4 RNA extraction and cDNA synthesis
The RNA of different varieties and different tissue samples were extracted by plant total RNA extraction kit (R4152) of Megan Biotechnology Co., Ltd. And the specific operation is shown in the instructions. The first strand cDNA was synthesized with total RNA as template, and reverse transcription was performed with HiScript III 1st Strand cDNA Synthesis Kit (+gDNA wiper) kit of Vazyme Biotech Co., Ltd. cDNA was synthesized according to the instructions of reverse transcriptase.
3.5 Primer design, gene cloning and sanger sequencing
Specific primers (Table 1) were designed based on the gene sequence information of the identified candidate RsPALs members to amplify the open reading frame in red radish ‘Hongxin No.1’ with high-fidelity enzyme (Takara PrimerSTAR Max DNA Polymerase). After electrophoretic detection, the amplified products were cut and recovered, and then the flat-end clone vector pTOPO-Blunt Simple (Aidlab Biotechnologies Co., Ltd.) was ligated and transformed into E. coli DH5α competent cells. After correcting monoclonal detection, the plasmid was extracted and sent to BGI for sequencing.
|
Table 1 primers used in this study |
3.6 Sequence alignment and phylogenetic tree analysis
The corresponding amino acid sequences were deduced by Translate Tool (http://cn.expasy.org/tools/dna.html) of ExPASy. The sequences were compared and analyzed by Clustal X software, and then the phylogenetic tree was constructed by MEGA5.0 to analyze the phylogenetic relationship (Tamura et al., 2011).
3.7 Fluorescence quantitative PCR
Primer information (Table 1) was used to carry out Real-Time PCR reaction on Roche LC 480 II Real-Time PCR System. The reaction conditions were as follows: 95℃ for 15 s, 56℃ for 15 s, 72℃ for 35 s, 40 cycles. The specificity of primers was analyzed by melting curve after PCR cycle. 2-ΔΔCT method was used to perform data (Schmittgen and Livak, 2008), and calculate the relative expression of genes. All the above experiments were repeated 3 times. H2O was used as the negative control in each experiment, and RsRPII gene was selected as the internal reference gene (Lim et al., 2016).
Authors’ contributions
LB and DLN designed and carried out the study. LB, CCF, WQ, and TZH performed the statistical analysis, and drafted the manuscript. DLN, and XJ participated in the design of the study and results analysis. DLN conceived of the project, directed the design of the study, data analysis, draft, and revision. All authors read and approved the final manuscript.
Acknowledgments
This study was supported by Special Foundation for Basic Science and Frontier Technology Research in Chongqing (cstc2017jcyjA0001), Science and Technology Research Project of Chongqing Education Commission (KJ1712303 and KJ1712305), and Research Project of Yangtze Normal University (2016KYQD19 and 2016KYQD20).
Chang Y.N., Liu J., and Liang J.T., 2018, Bioinformatics analysis of PAL gene in potato, Xiandai Nongye Yanjiu, 30(6): 5-8
Chen M., Zhang X., Zhang Y., Yang M.H., and Liu H.F., 2018, Cloning and expression analysis of phenylalanine (PAL) gene in Vitis amurensis, Huabei Nongxuebao (ACTA Agriculturae Borealio-Sinica), 33(6): 64-71
Deng L.C., Cui L.N., Yang L., Chen J., He W.Z., Li X., Tang H.T., Zhang B., Kang J.W., Yang J.P., and Tan J., 2019, Identification of gene family of Phenylalanine Ammonia-Lyase and analysis of resistance to maize sheath blight in corn, Fenzi Zhiwu Yuzhong (Molecular Plant Breeding), 17(3): 891-897
Feng L.J., Yin Y.L., Jiao Q.Q., and Yang X.M., 2018, Cloning and expression analysis of PAL gene in pomegranate (Punica granatum L.), Henongxue Bao (Journal of Nuclear Agricultural Sciences), 32(7): 1320-1329
Hao X.Y., Sun X.L., Wang T.C., Lü K.L., Lai Z.X. and Cheng C.Z., 2018, Characteristics and functions of plant Phenylalanine Ammonia Lyase genes and the encoded proteins, Redai Zuowu Xuebao (Chinese Journal of Tropical Crops), 39(7): 1452-1461
Huang J.L., Gu M., Lai Z.B., Fan B.F., Shi K., Zhou Y.Z., Yu J.Q., and Chen Z.X., 2010, Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress, Plant Physiol., 153(4): 1526-1538
https://doi.org/10.1104/pp.110.157370
PMid:20566705 PMCid:PMC2923909
Koukol J., Miljanich P., and Conn E.E., 1962, The metabolism of aromatic compounds in higher plants, J. Biol. Chem., 237(10): 2692-2698
https://doi.org/10.1016/S0021-9258(19)61721-7
Li J.S., Yang R., Sui X.S., Cheng J.H., Cao Q.Q., and Wang S.H., 2008, Analysis and evaluation on nutritive quality indicators of different genotypes radish, Huabei Nongxuebao (ACTA Agriculturae Borealio-Sinica), 23(S1): 77-80
Lim S., Song J., Kim D., Kim J. K., Lee J., Kim Y., and Ha S., 2016, Activation of anthocyanin biosynthesis by expression of the radish R2R3-MYB transcription factor gene RsMYB1, Plant Cell Reports, 35(3): 641-653
https://doi.org/10.1007/s00299-015-1909-3
PMid:26703384
Muleke E.M., Fan L., Wang Y., Xu L., Zhu X., Zhang W., Cao Y., Karanja B.K., and Liu L., 2017, Coordinated regulation of anthocyanin biosynthesis genes confers varied phenotypic and Spatial-Temporal anthocyanin accumulation in radish (Raphanus sativus L.), Front. Plant Sci., 8: 1243
https://doi.org/10.3389/fpls.2017.01243
PMid:28769952 PMCid:PMC5515825
Olsen K.M., Lea U.S., Slimestad R., Verheul M. and Lillo C., 2008, Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis, J. Plant Physiol., 165(14): 1491-1499
https://doi.org/10.1016/j.jplph.2007.11.005
PMid:18242769
Pombo M.A., Martínez G.A., and Civello P.M., 2011, Cloning of FaPAL6 gene from strawberry fruit and characterization of its expression and enzymatic activity in two cultivars with different anthocyanin accumulation, Plant Sci., 181(2): 111-118
https://doi.org/10.1016/j.plantsci.2011.04.012
PMid:21683875
Schmittgen T.D., and Livak K.J., 2008, Analyzing real-time PCR data by the comparative C(T) method, Nat. Protoc., 3(6): 1101-1108
https://doi.org/10.1038/nprot.2008.73
PMid:18546601
Su N., Lu Y.W., Wu Q., Liu Y.Y., Xia Y., Xia K., and Cui J., 2015, UV-B-induced anthocyanin accumulation in hypocotyls of radish sprouts continues in the dark after irradiation, J. Sci. Food Agric., 96(3): 886-892
https://doi.org/10.1002/jsfa.7161
PMid:25754879
Sun Y.J., Chen X., Cui H.N., Wu C., Liu S., Luan F.S., and Wang X.Z., 2018, Bioinformatics analysis of PAL gene family in cucurbitaceae and the PAL4 gene cloning in Cucumis melo L., Fenzi Zhiwu Yuzhong (Molecular Plant Breeding), 16(15): 4910-4920
Tamura K., Peterson D., Peterson N., Stecher G., Nei M., and Kumar S., 2011, MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods, Mol. Biol. Evol., 28(10): 2731-2739
https://doi.org/10.1093/molbev/msr121
PMid:21546353 PMCid:PMC3203626
Winkel-Shirley B., 2001, Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology, Plant Physiol., 126(2): 485-493
https://doi.org/10.1104/pp.126.2.485
PMid:11402179 PMCid:PMC1540115
Wrolstad R.E., Culbertson J.D., Cornwell C.J., and Mattick L.R., 1982, Detection of adulteration in blackberry juice concentrates and wines, J. Assoc. Off. Anal. Chem., 65(6): 1417-1423
https://doi.org/10.1093/jaoac/65.6.1417
Xiong F., Lu Q.H., Fang W., Yang Y., Wang X.C., Zhu X.J. and Wang Y.C., 2020, Genome-wide identification and expression analyses of PAL genes under biotic and abiotic stress in Camellia sinensis, Yuanyi Xuebao (Acta Horticulturae Sinica), 47(3): 517-528
Xu C.M., Pang G.Y., and Li T., 2013, Progress in the research on physiological activities of anthocyanin, Zhongguo Shipin Tianjiaji (China Food Additives), (3): 205-210
Yang G.D., Duan L., Shu Q.L., and Huang C.C., 2007, Relationship of anthocyanidin content, sugar content, PAL activity and Colletotrichum gloeosporioides in peel of oil tea tree, Linye Kexue (Scientia Silvae Sinicae), 43(6): 100-104
Yang H.X., Sun Y.Y., Jia C.H., Jin Z.Q., Xu B.Y., and Wang Z., 2019, Identification and expression analysis of phenylalanine ammonialyase gene family in banana, 40(10): 1949-1957
Zeng J.L., Ouyang L.J., Liu J.L., He H.H., Zhu C.L., Peng X.S., He X.P., Fu J.R., Chen X.R., Bian J.M., Xu J., Sun X.T., Zhou D.H., and Hu L.F., 2018, Whole genome analysis and stress expression research of PAL gene in rice, Jiyinzuxue Yu Yingyong Shengwuxue (Genomics and Applied Biology), 37(9): 3881-3888
Zhang L.Z., Fan S., An N., Zuo X.Y., Gao C., Zhang D., and Han M.Y., Identification and expression analysis of PAL gene family in apple, Zhejiang Nongye Xuebao (Acta Agriculturae Zhejiangensis), 30(12): 2031-2043
Zhang Z.C., Sun C.Q., Yao Y.M., Mao Z.L., Sun G.S., and Dai Z.L., 2019, Red anthocyanins contents and the relationships with phenylalanine ammonia lyase (PAL) activity, soluble sugar and chlorophyll contents in carmine radish (Raphanus sativus L.), Hortic. Sci., 46(1): 17-25
https://doi.org/10.17221/202/2017-HORTSCI
Zhou A.Q., Zhu J., Sheng J.P., Shen L., and Sheng Z.J., 1997, The relationship of anthocyanin formation, PAL activity and protein content during apple colouring, Zhongguo Nongye Daxue Xuebao (Journal of China Agricultural University), 2(3): 97-99
Zhu H.S., Wen W.X., Liu J.T., Ye R.X., Chen M.D., Wang B., Zhang Q.R., Li Y.P., and Wen Q.F., 2018, Cloning and expression analysis of Phenylalanine Ammonia-Lyase PAL gene from Luffa cylindrical, 19(2): 305-313
. PDF(1280KB)
. HTML
Associated material
. Readers' comments
Other articles by authors
. Biao Lai
. Chunfan Chen
. Zehui Tang
. Jing Xiao
. Qi Wang
. Fabo Chen
. Lina Du
Related articles
. Carmine radish
. PAL
. Anthocyanin
. Gene expression
Tools
. Email to a friend
. Post a comment
.png)
.png)
.png)

.png)


